4. dev with dask#
[1]:
from collections import defaultdict
import dask.array as da
import holoviews as hv
import hvplot
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import skimage
import tifffile as tff
from scipy import ndimage
from nima import nima, utils
%load_ext autoreload
%autoreload 2
fp = "../../tests/data/1b_c16_15.tif"
[2]:
daimg = da.from_zarr(tff.imread(fp, aszarr=True))
daimg
[2]:
|
||||||||||||||||
[3]:
utils.bg(daimg[0, 0].compute())
[3]:
(np.float64(457.8380994040984), np.float64(48.50287340564226))
[4]:
def dabg(daimg):
r = defaultdict(list)
n_t, n_c = daimg.shape[:2]
for t in range(n_t):
for c in range(n_c):
r[c].append(utils.bg(daimg[t, c].compute())[0])
return pd.DataFrame(r)
dabg(daimg)
[4]:
| 0 | 1 | 2 | |
|---|---|---|---|
| 0 | 457.838099 | 257.010244 | 289.378226 |
| 1 | 457.295254 | 259.072941 | 289.627118 |
| 2 | 457.760167 | 260.182049 | 290.268666 |
| 3 | 453.995203 | 257.189940 | 285.613624 |
[5]:
def dabg_fg(daimg, erf_pvalue=1e-100, size=10):
n_t, n_c = daimg.shape[:2]
bgs = defaultdict(list)
fgs = defaultdict(list)
for t in range(n_t):
p = np.ones(daimg.shape[-2:])
multichannel = daimg[t].compute()
for c in range(n_c):
av, sd = utils.bg(multichannel[c])
p = p * utils.prob(multichannel[c], av, sd)
bgs[c].append(av)
mask = ndimage.median_filter((p) ** (1 / n_c), size=size) < erf_pvalue
for c in range(n_c):
fgs[c].append(np.ma.mean(np.ma.masked_array(multichannel[c], mask=~mask)))
return pd.DataFrame(bgs), pd.DataFrame(fgs)
dfb, dff = dabg_fg(daimg)
[6]:
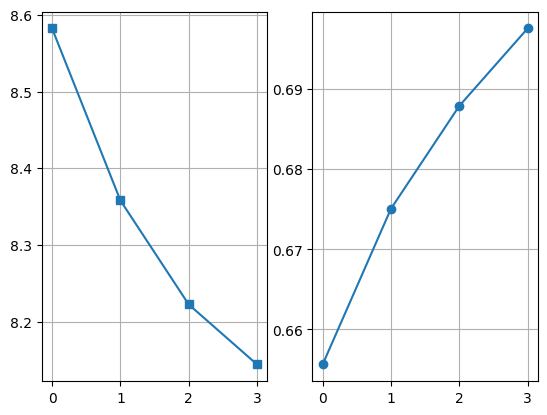
plt.subplot(121)
((dff - dfb)[0] / (dff - dfb)[2]).plot(marker="s")
plt.grid()
plt.subplot(122)
((dff - dfb)[2] / (dff - dfb)[1]).plot(marker="o")
plt.grid()
NEXT:
make utils.bg and utils.prob work with dask arrays
[7]:
def dmask(daim, erf_pvalue=1e-100, size=10):
n_c = daim.shape[0]
im = daim[0].compute()
p = utils.prob(im, *utils.bg(im))
for c in range(1, n_c):
im = daim[c].compute()
p = p * utils.prob(im, *utils.bg(im))
p = ndimage.median_filter((p) ** (1 / n_c), size=size)
mask = p < erf_pvalue
return skimage.morphology.remove_small_objects(mask)
# mask = skimage.morphology.remove_small_holes(mask)
# return np.ma.masked_array(plane, mask=~mask), np.ma.masked_array(plane, mask=mask)
mask = dmask(daimg[2])
lab, nlab = ndimage.label(mask)
lab, nlab
[7]:
(array([[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0]], shape=(512, 512), dtype=int32),
2)
[8]:
pr = skimage.measure.regionprops(lab, intensity_image=daimg[0][0])
pr[1].equivalent_diameter
[8]:
np.float64(195.49311541527658)
[9]:
max_diameter = pr[0].equivalent_diameter
size = int(max_diameter * 0.3)
size
[9]:
47
[10]:
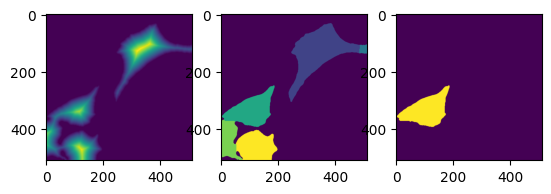
t = 0
mask = dmask(daimg[t])
# skimage.io.imshow(mask)
lab, nlab = ndimage.label(mask)
distance = ndimage.distance_transform_edt(mask)
# distance = skimage.filters.gaussian(distance, sigma=0) min_distance=size,
coords = skimage.feature.peak_local_max(
distance, footprint=np.ones((size, size)), labels=lab
)
mm = np.zeros(distance.shape, dtype=bool)
mm[tuple(coords.T)] = True
# markers, _ = ndimage.label(mm)
markers = skimage.measure.label(mm)
labels = skimage.segmentation.watershed(-distance, markers, mask=mask)
_, (ax0, ax1, ax2) = plt.subplots(1, 3)
ax0.imshow(distance)
ax1.imshow(labels)
ax2.imshow(labels == 3)
coords
[10]:
array([[122, 329],
[122, 510],
[475, 125],
[341, 116],
[421, 1]])
[11]:
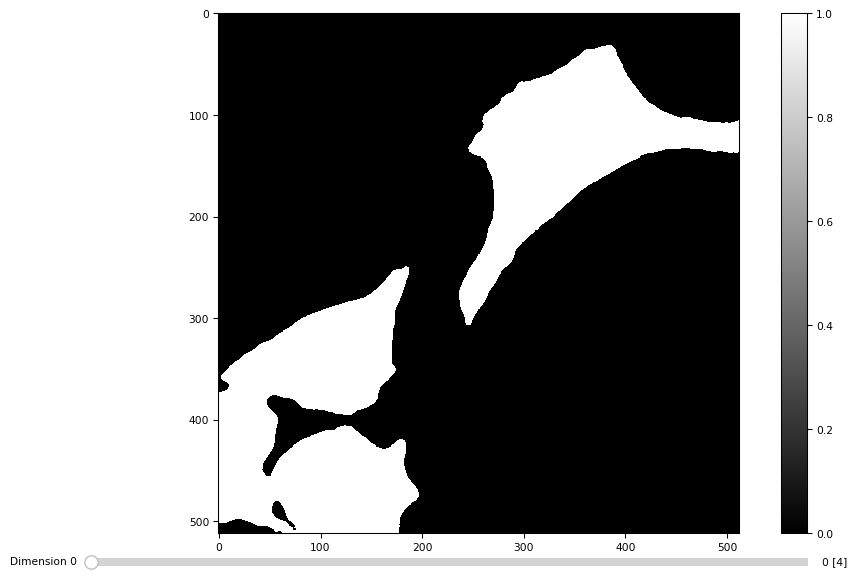
masks = [dmask(daimg[t]) for t in range(4)]
[12]:
masks = np.stack(masks)
masks.shape
[12]:
(4, 512, 512)
[13]:
tff.imshow(masks)
[13]:
(<Figure size 988.8x604.8 with 3 Axes>,
<Axes: >,
<matplotlib.image.AxesImage at 0x7f5294a87750>)
[14]:
distance = ndimage.distance_transform_edt(masks)
distance = skimage.filters.gaussian(distance, sigma=5)
[15]:
import impy
impy.array(distance).imshow()
[15]:
| name | No name |
| shape | 4(t), 512(y), 512(x) |
| label shape | No label |
| dtype | float64 |
| source | None |
| scale | ScaleView(t=1.0000, y=1.0000, x=1.0000) |
[16]:
for t in range(4):
coords = skimage.feature.peak_local_max(distance[t], footprint=np.ones((130, 130)))
print(coords)
[[114 346]
[473 128]
[344 110]]
[[114 346]
[473 128]
[344 110]]
[[114 346]
[473 128]
[344 110]]
[[114 346]
[473 128]
[344 110]]
[17]:
co = np.stack([coords, coords, coords, coords])
[18]:
coords.T
[18]:
array([[114, 473, 344],
[346, 128, 110]])
[19]:
mm = np.zeros(masks[0].shape, dtype=bool)
mm[tuple(co.T)] = True
# markers, _ = ndimage.label(mm)
markers = skimage.measure.label(np.stack([mm, mm, mm, mm]))
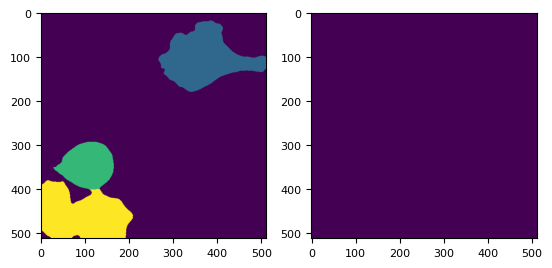
labels = skimage.segmentation.watershed(-distance, markers, mask=masks)
_, (ax1, ax2) = plt.subplots(1, 2)
ax1.imshow(labels[3])
ax2.imshow(labels[3] == 4)
[19]:
<matplotlib.image.AxesImage at 0x7f5294354410>
[20]:
img = tff.imread(fp)
[21]:
dim, _, _ = nima.read_tiff(fp, channels=["R", "G", "C"])
(4, 512, 512)
[22]:
res = nima.d_bg(dim)
bgs = res[1]
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[22], line 1
----> 1 res = nima.d_bg(dim)
2 bgs = res[1]
TypeError: d_bg() missing 1 required positional argument: 'bg_params'
[23]:
def ratio(t, roi):
g = img[t, 0][labels[t] == roi].mean() - bgs["G"][t]
r = img[t, 1][labels[t] == roi].mean() - bgs["R"][t]
c = img[t, 2][labels[t] == roi].mean() - bgs["C"][t]
return g / c, c / r
ratio(1, 4)
/tmp/ipykernel_2191/533071019.py:2: RuntimeWarning: Mean of empty slice.
g = img[t, 0][labels[t] == roi].mean() - bgs["G"][t]
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/numpy/_core/_methods.py:144: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[23], line 8
4 c = img[t, 2][labels[t] == roi].mean() - bgs["C"][t]
5 return g / c, c / r
----> 8 ratio(1, 4)
Cell In[23], line 2, in ratio(t, roi)
1 def ratio(t, roi):
----> 2 g = img[t, 0][labels[t] == roi].mean() - bgs["G"][t]
3 r = img[t, 1][labels[t] == roi].mean() - bgs["R"][t]
4 c = img[t, 2][labels[t] == roi].mean() - bgs["C"][t]
NameError: name 'bgs' is not defined
[24]:
rph = defaultdict(list)
rcl = defaultdict(list)
for roi in range(1, 5):
for t in range(4):
ph, cl = ratio(t, roi)
rph[roi].append(ph)
rcl[roi].append(cl)
plt.plot(rph[1])
plt.plot(rph[2])
plt.plot(rph[3])
plt.plot(rph[4])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[24], line 5
3 for roi in range(1, 5):
4 for t in range(4):
----> 5 ph, cl = ratio(t, roi)
6 rph[roi].append(ph)
7 rcl[roi].append(cl)
Cell In[23], line 2, in ratio(t, roi)
1 def ratio(t, roi):
----> 2 g = img[t, 0][labels[t] == roi].mean() - bgs["G"][t]
3 r = img[t, 1][labels[t] == roi].mean() - bgs["R"][t]
4 c = img[t, 2][labels[t] == roi].mean() - bgs["C"][t]
NameError: name 'bgs' is not defined
[25]:
plt.plot(rcl[1])
plt.plot(rcl[2])
plt.plot(rcl[3])
plt.plot(rcl[4])
[25]:
[<matplotlib.lines.Line2D at 0x7f528cb35f90>]
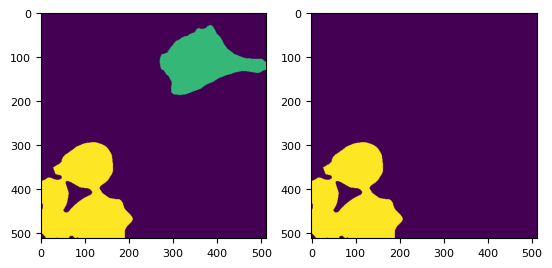
[26]:
t = 2
mask = dmask(daimg[t])
# skimage.io.imshow(mask)
lab, nlab = ndimage.label(mask)
lab[~mask] = -1
# lab[lab==1] = -1
labels_ws = skimage.segmentation.random_walker(
daimg[t, 1].compute(), lab, beta=1e10, mode="bf"
)
# labels_ws = skimage.segmentation.random_walker(-distance, lab, beta=10000, mode="bf")
_, (ax1, ax2) = plt.subplots(1, 2)
ax1.imshow(labels_ws)
ax2.imshow(labels_ws == 2)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/segmentation/random_walker_segmentation.py:545: UserWarning: Random walker only segments unlabeled areas, where labels == 0. No zero valued areas in labels were found. Returning provided labels.
(labels, nlabels, mask, inds_isolated_seeds, isolated_values) = _preprocess(labels)
[26]:
<matplotlib.image.AxesImage at 0x7f528cbe20d0>
[27]:
imar = impy.imread(fp)
imar.label_threshold()
[27]:
| name | 1b_c16_15.tif |
| shape | 4(t), 3(c), 512(y), 512(x) |
| dtype | uint16 |
| source | ../../tests/data/1b_c16_15.tif |
| scale | ScaleView(t=1.0000, c=1.0000, y=0.2000, x=0.2000) |
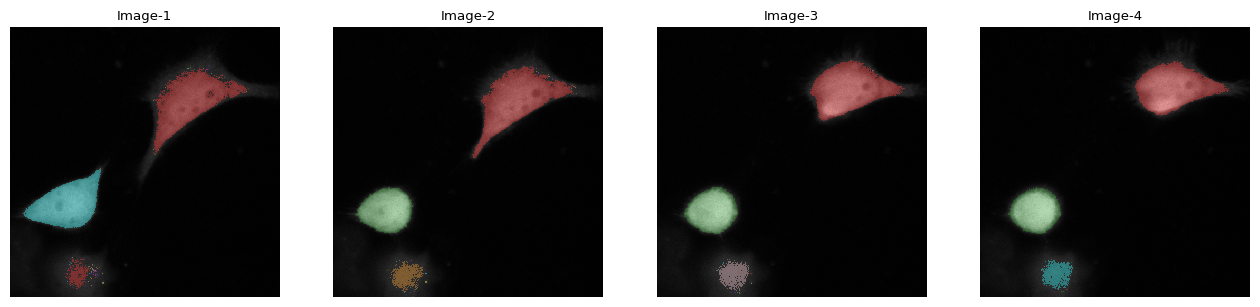
[28]:
imar[:, 2].imshow(label=1)
[28]:
| name | 1b_c16_15.tif |
| shape | 4(t), 512(y), 512(x) |
| label shape | 4(t), 512(y), 512(x) |
| dtype | uint16 |
| source | ../../tests/data/1b_c16_15.tif |
| scale | ScaleView(t=1.0000, y=0.2000, x=0.2000) |
[29]:
def dmask0(im, erf_pvalue=1e-100, size=10):
p = utils.prob(im[0], *utils.bg(im[0]))
for img in im[1:]:
p = p * utils.prob(img, *utils.bg(img))
p = ndimage.median_filter((p) ** (1 / len(im)), size=size)
mask = p < erf_pvalue
return skimage.morphology.remove_small_objects(mask)
[30]:
dmask0(imar[1])
[30]:
array([[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]], shape=(512, 512))
[31]:
plt.imshow(skimage.measure.label(mask))
[31]:
<matplotlib.image.AxesImage at 0x7f528c908910>
[32]:
distance = skimage.filters.gaussian(distance, sigma=30)
tff.imshow(distance)
[32]:
(<Figure size 988.8x604.8 with 1 Axes>,
<Axes: >,
<matplotlib.image.AxesImage at 0x7f528c766990>)
[33]:
np.transpose(np.nonzero(skimage.morphology.local_maxima(distance)))
[33]:
array([[ 3, 110, 353],
[ 3, 346, 107],
[ 3, 456, 18],
[ 3, 486, 128]])
[34]:
tff.imshow(ndimage.label(mask)[0])
[34]:
(<Figure size 988.8x604.8 with 2 Axes>,
<Axes: >,
<matplotlib.image.AxesImage at 0x7f528c7e8910>)
[35]:
res[1]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[35], line 1
----> 1 res[1]
NameError: name 'res' is not defined
[36]:
res[2]["G"][2][0]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[36], line 1
----> 1 res[2]["G"][2][0]
NameError: name 'res' is not defined
[37]:
res[1].plot()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[37], line 1
----> 1 res[1].plot()
NameError: name 'res' is not defined
[38]:
import hvplot.pandas
[39]:
res[1].hvplot()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[39], line 1
----> 1 res[1].hvplot()
NameError: name 'res' is not defined
[40]:
import xarray as xr
[41]:
xim = xr.DataArray(
data=[dim["G"], dim["R"], dim["C"]],
dims=["channel", "time", "y", "x"],
coords={
"channel": ["Green", "Red", "Cyan"],
"time": [0, 1, 2, 3],
"y": range(512),
"x": range(512),
},
)
[42]:
import hvplot.xarray
[43]:
xim.sel(time=0, channel="Green").hvplot(width=400, height=300)
[43]:
[44]:
xim.sel(time=0).hvplot(
width=300,
subplots=True,
by="channel",
)
[44]:
[45]:
hvplot.extension(
"bokeh",
"matplotlib",
)
[46]:
img = xim.sel(time=0).sel(channel="Red")
[47]:
hvimg = hv.Image(img)
[48]:
# %%opts Image [aspect=1388/1038]
f = xim.sel(channel="Red").hvplot(
frame_width=300,
frame_height=200,
subplots=True,
col="time",
yaxis=False,
colorbar=False,
xaxis=False,
cmap="Reds",
) + xim.sel(channel="Cyan").hvplot(
subplots=True, col="time", yaxis=False, colorbar=False, xaxis=False, cmap="Greens"
)
f
[48]:
[49]:
import aicsimageio
aicsimageio.__version__
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[49], line 1
----> 1 import aicsimageio
3 aicsimageio.__version__
ModuleNotFoundError: No module named 'aicsimageio'
[50]:
reader = aicsimageio.readers.tiff_reader.TiffReader
aim1 = aicsimageio.AICSImage(
"/home/dati/dt-evolv/data/2022-06-17/images/Vero-Hek/2022-06-14/13080/TimePoint_1/6w-20Xph1-SpikeTest3_A02_s570_w14510D534-71A3-4EB5-B48F-F4331FE96517.tif",
reader=reader,
)
aim2 = aicsimageio.AICSImage(
"/home/dati/dt-evolv/data/2022-06-17/images/Vero-Hek/2022-06-14/13080/TimePoint_1/6w-20Xph1-SpikeTest3_A02_s570_w25049D5AC-5888-492F-891D-8BECC1AB67DF.tif",
reader=reader,
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[50], line 1
----> 1 reader = aicsimageio.readers.tiff_reader.TiffReader
2 aim1 = aicsimageio.AICSImage(
3 "/home/dati/dt-evolv/data/2022-06-17/images/Vero-Hek/2022-06-14/13080/TimePoint_1/6w-20Xph1-SpikeTest3_A02_s570_w14510D534-71A3-4EB5-B48F-F4331FE96517.tif",
4 reader=reader,
5 )
6 aim2 = aicsimageio.AICSImage(
7 "/home/dati/dt-evolv/data/2022-06-17/images/Vero-Hek/2022-06-14/13080/TimePoint_1/6w-20Xph1-SpikeTest3_A02_s570_w25049D5AC-5888-492F-891D-8BECC1AB67DF.tif",
8 reader=reader,
9 )
NameError: name 'aicsimageio' is not defined
[51]:
x1 = aim1.xarray_data
x2 = aim2.xarray_data
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[51], line 1
----> 1 x1 = aim1.xarray_data
2 x2 = aim2.xarray_data
NameError: name 'aim1' is not defined
[52]:
# Create a new Dataset with new coordinates
ds = xr.Dataset({"c1": x1, "c2": x2})
# Assuming ds is your Dataset
new_coords = {"Frame": [1, 2], "excitation_wavelength": [400, 500]}
# Use assign_coords to set new coordinates
ds_assigned = ds.assign_coords(**new_coords)
ds_assigned
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[52], line 2
1 # Create a new Dataset with new coordinates
----> 2 ds = xr.Dataset({"c1": x1, "c2": x2})
4 # Assuming ds is your Dataset
5 new_coords = {"Frame": [1, 2], "excitation_wavelength": [400, 500]}
NameError: name 'x1' is not defined
[53]:
aim2.metadata[220:230] == aim1.metadata[220:230]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[53], line 1
----> 1 aim2.metadata[220:230] == aim1.metadata[220:230]
NameError: name 'aim2' is not defined
[54]:
im = x1.to_numpy()[0, 0, 0]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[54], line 1
----> 1 im = x1.to_numpy()[0, 0, 0]
NameError: name 'x1' is not defined
[55]:
im1 = tff.imread("/home/dati/dt-evolv/data/2022-06-17/flat_w1.tif")
im2 = tff.imread("/home/dati/dt-evolv/data/2022-06-17/flat_w2.tif")
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2191/2721790190.py in ?()
----> 1 im1 = tff.imread("/home/dati/dt-evolv/data/2022-06-17/flat_w1.tif")
2 im2 = tff.imread("/home/dati/dt-evolv/data/2022-06-17/flat_w2.tif")
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(files, selection, aszarr, key, series, level, squeeze, maxworkers, buffersize, mode, name, offset, size, pattern, axesorder, categories, imread, imreadargs, sort, container, chunkshape, chunkdtype, axestiled, ioworkers, chunkmode, fillvalue, zattrs, multiscales, omexml, superres, out, out_inplace, _multifile, _useframes, **kwargs)
1172 ):
1173 files = files[0]
1174
1175 if isinstance(files, str) or not isinstance(files, Sequence):
-> 1176 with TiffFile(
1177 files,
1178 mode=mode,
1179 name=name,
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-evolv/data/2022-06-17/flat_w1.tif'
[56]:
%%opts Image [aspect=1388/1038]
%%opts Image.Cyan style(cmap=plt.cm.Blues)
%%opts Image.Green style(cmap=plt.cm.Greens)
%%opts Image.Red style(cmap=plt.cm.Reds)
/tmp/ipykernel_2191/1736577656.py:1: HoloviewsDeprecationWarning: IPython magic is deprecated and will be removed in version 1.23.0.
get_ipython().run_cell_magic('opts', 'Image [aspect=1388/1038]', '\n%%opts Image.Cyan style(cmap=plt.cm.Blues)\n%%opts Image.Green style(cmap=plt.cm.Greens)\n%%opts Image.Red style(cmap=plt.cm.Reds)\n')
[57]:
chans = (
hv.Image(dim["C"][0], group="cyan")
+ hv.Image(dim["G"][2], group="green")
+ hv.Image(dim["R"][1], group="red")
)
chans
[57]:
[58]:
hv.save(chans, "a.png")
5. Holoviews#
[59]:
hv.notebook_extension()
cm = plt.cm.inferno_r
channels = ["G", "R", "C"]
dim, n_ch, times = nima.read_tiff(fp, channels)
dimm = nima.d_median(dim)
f = nima.d_show(dimm, cmap=cm)
/tmp/ipykernel_2191/1764924339.py:1: HoloviewsDeprecationWarning: Calling 'hv.extension()' without arguments is deprecated and will be removed in version 1.23.0, use 'hv.extension("matplotlib")' instead.
hv.notebook_extension()
(4, 512, 512)
[60]:
%%opts Image [aspect=512/512]
%%opts Image.Cyan style(cmap=plt.cm.Blues)
%%opts Image.Green style(cmap=plt.cm.Greens)
%%opts Image.Red style(cmap=plt.cm.Reds)
chans = hv.Image(dim['C'][0], group='cyan') \
+ hv.Image(dim['G'][0], group='green') \
+ hv.Image(dim['R'][0], group='red')
chans
/tmp/ipykernel_2191/3523771648.py:1: HoloviewsDeprecationWarning: IPython magic is deprecated and will be removed in version 1.23.0.
get_ipython().run_cell_magic('opts', 'Image [aspect=512/512]', "\n%%opts Image.Cyan style(cmap=plt.cm.Blues)\n%%opts Image.Green style(cmap=plt.cm.Greens)\n%%opts Image.Red style(cmap=plt.cm.Reds)\n\nchans = hv.Image(dim['C'][0], group='cyan') \\\n + hv.Image(dim['G'][0], group='green') \\\n + hv.Image(dim['R'][0], group='red')\n\nchans\n")
[60]:
[61]:
c = [(i, hv.Image(im)) for i, im in enumerate(dim["C"])]
c = hv.HoloMap(c, kdims=["Frame"])
g = [(i, hv.Image(im)) for i, im in enumerate(dim["G"])]
g = hv.HoloMap(g, kdims=["Frame"])
r = [(i, hv.Image(im)) for i, im in enumerate(dim["R"])]
r = hv.HoloMap(r, kdims=["Frame"])
[62]:
%%output holomap='auto'
%%opts Image style(cmap='viridis')
(c + g).select(Frame={0,5,6,7,10,30}).cols(2)
/tmp/ipykernel_2191/3640934118.py:1: HoloviewsDeprecationWarning: IPython magic is deprecated and will be removed in version 1.23.0.
get_ipython().run_cell_magic('output', "holomap='auto'", "%%opts Image style(cmap='viridis')\n(c + g).select(Frame={0,5,6,7,10,30}).cols(2)\n")
/tmp/ipykernel_2191/1261574846.py:1: HoloviewsDeprecationWarning: IPython magic is deprecated and will be removed in version 1.23.0.
get_ipython().run_cell_magic('opts', "Image style(cmap='viridis')", '(c + g).select(Frame={0,5,6,7,10,30}).cols(2)\n')
[62]:
[63]:
c[::20].overlay("Frame")
[63]:
[64]:
wl = hv.Dimension("excitation wavelength", unit="nm")
c = c.add_dimension(wl, 1, 458)
g = g.add_dimension(wl, 1, 488)
r = r.add_dimension(wl, 1, 561)
channels = c.clone()
channels.update(g)
channels.update(r)
[65]:
%%opts Image style(cmap='viridis')
%%output size=300
channels[::5].grid(['Frame', 'excitation wavelength'])
/tmp/ipykernel_2191/2279777199.py:1: HoloviewsDeprecationWarning: IPython magic is deprecated and will be removed in version 1.23.0.
get_ipython().run_cell_magic('opts', "Image style(cmap='viridis')", "%%output size=300\nchannels[::5].grid(['Frame', 'excitation wavelength'])\n")
/tmp/ipykernel_2191/3322840880.py:1: HoloviewsDeprecationWarning: IPython magic is deprecated and will be removed in version 1.23.0.
get_ipython().run_cell_magic('output', 'size=300', "channels[::5].grid(['Frame', 'excitation wavelength'])\n")
[65]:
[66]:
t = [(i, hv.Image(im)) for i, im in enumerate(dim["C"])]
[67]:
hv.HoloMap([(i, hv.Image(im)) for i, im in enumerate(dim["C"])], kdims=["frame"])
[67]:
[68]:
hv.NdLayout(
{
k: hv.HoloMap(
[(i, hv.Image(im)) for i, im in enumerate(dim[k])], kdims=["frame"]
)
for k in dim
},
kdims=["channels"],
)[::4]
[68]:
[69]:
%%opts Image (cmap='viridis')
%%opts Image.A [aspect=2]
im = hv.Image(dim["G"][1], bounds=(0, 0, 512, 512))
im2 = hv.Image(dim['C'][1], bounds=(0, 0, 512, 512))
im3 = hv.Image(dimm['C'][1], bounds=(0, 0, 512, 512))
((im * hv.HLine(y=350)) + im.sample(y=350) + (im2 * hv.HLine(y=150)) + im2.sample(y=150) * im3.sample(y=150)).cols(3)
/tmp/ipykernel_2191/4117421567.py:1: HoloviewsDeprecationWarning: IPython magic is deprecated and will be removed in version 1.23.0.
get_ipython().run_cell_magic('opts', "Image (cmap='viridis')", '%%opts Image.A [aspect=2]\nim = hv.Image(dim["G"][1], bounds=(0, 0, 512, 512))\nim2 = hv.Image(dim[\'C\'][1], bounds=(0, 0, 512, 512))\nim3 = hv.Image(dimm[\'C\'][1], bounds=(0, 0, 512, 512))\n((im * hv.HLine(y=350)) + im.sample(y=350) + (im2 * hv.HLine(y=150)) + im2.sample(y=150) * im3.sample(y=150)).cols(3)\n')
[69]: