3. Image simulation with dask and generat#
The purpose here is to simulate images to identify the best methods for:
Determining the FLAT image
Segmenting cells from the background
Computing the ratio
Determine the minimal detectable gradient for a given error.
Since subtracting the correct background value is crucial for accurate ratio imaging, we will test the distribution of background values with probplot for normality.
External examples: https://cbia.fi.muni.cz/simulator/
[1]:
%load_ext autoreload
%autoreload 2
import dask.array as da
import dask_image
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import scipy
import skimage
import skimage.filters
import skimage.io
import tifffile as tff
import zarr
from dask_image import ndfilters, ndmorph
from scipy import ndimage
from nima import generat, nima, segmentation, utils
nrg = np.random.default_rng(1)
3.1. FlatXY#
[2]:
fp_flatxy = "/home/dati/dt-clop3/data/210920/flatxy.tf8"
[3]:
nima.read_tiffmd(fp_flatxy, channels=["G", "R", "C"])
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2378/4002580391.py in ?()
----> 1 nima.read_tiffmd(fp_flatxy, channels=["G", "R", "C"])
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
1072 def read_tiffmd(fp: Path, channels: Sequence[str]) -> tuple[Array, Metadata]:
1073 """Read multichannel TIFF timelapse image."""
1074 n_channels = len(channels)
-> 1075 rdr = TiffReader(fp)
1076 dim = da.from_zarr(rdr.aszarr()) # type: ignore[no-untyped-call]
1077 md = Metadata(rdr)
1078 if md.size_c[0] % n_channels:
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/210920/flatxy.tf8'
[4]:
store = tff.imread(fp_flatxy, aszarr=True)
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2378/1232588744.py in ?()
----> 1 store = tff.imread(fp_flatxy, aszarr=True)
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(files, selection, aszarr, key, series, level, squeeze, maxworkers, buffersize, mode, name, offset, size, pattern, axesorder, categories, imread, imreadargs, sort, container, chunkshape, chunkdtype, axestiled, ioworkers, chunkmode, fillvalue, zattrs, multiscales, omexml, superres, out, out_inplace, _multifile, _useframes, **kwargs)
1172 ):
1173 files = files[0]
1174
1175 if isinstance(files, str) or not isinstance(files, Sequence):
-> 1176 with TiffFile(
1177 files,
1178 mode=mode,
1179 name=name,
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/210920/flatxy.tf8'
[5]:
zc1a = zarr.open(store)
zc1a.info
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[5], line 1
----> 1 zc1a = zarr.open(store)
2 zc1a.info
NameError: name 'store' is not defined
[6]:
dd = da.from_zarr(store)
dd
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[6], line 1
----> 1 dd = da.from_zarr(store)
2 dd
NameError: name 'store' is not defined
[7]:
img = dd[0, 0]
plt.imshow(img, vmax=60)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[7], line 1
----> 1 img = dd[0, 0]
2 plt.imshow(img, vmax=60)
NameError: name 'dd' is not defined
[8]:
from dask.diagnostics.progress import ProgressBar
with ProgressBar():
bg_result = nima.bg(img.compute())
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[8], line 4
1 from dask.diagnostics.progress import ProgressBar
3 with ProgressBar():
----> 4 bg_result = nima.bg(img.compute())
AttributeError: module 'nima.nima' has no attribute 'bg'
[9]:
bg_result.figures[1]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[9], line 1
----> 1 bg_result.figures[1]
NameError: name 'bg_result' is not defined
[10]:
im = img.compute()
bgmax = segmentation._bgmax(im, bins=25, densityplot=True)
bgmax
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[10], line 1
----> 1 im = img.compute()
2 bgmax = segmentation._bgmax(im, bins=25, densityplot=True)
3 bgmax
NameError: name 'img' is not defined
[11]:
bg_result = segmentation.bg_refine_iteratively(im, bgmax=bgmax, probplot=1)
bg, sd, iqr, figs = bg_result.bg, bg_result.sd, bg_result.iqr, bg_result.figures
bg, sd, iqr, figs
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[11], line 1
----> 1 bg_result = segmentation.bg_refine_iteratively(im, bgmax=bgmax, probplot=1)
2 bg, sd, iqr, figs = bg_result.bg, bg_result.sd, bg_result.iqr, bg_result.figures
3 bg, sd, iqr, figs
AttributeError: module 'nima.segmentation' has no attribute 'bg_refine_iteratively'
[12]:
pp = da.mean(
dask_image.ndfilters.maximum_filter(dd[0:4000:20, 0], size=(100, 1, 1)), axis=0
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[12], line 2
1 pp = da.mean(
----> 2 dask_image.ndfilters.maximum_filter(dd[0:4000:20, 0], size=(100, 1, 1)), axis=0
3 )
NameError: name 'dd' is not defined
[13]:
ppp = pp.compute()
plt.imshow(skimage.filters.gaussian(ppp, 100))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[13], line 1
----> 1 ppp = pp.compute()
3 plt.imshow(skimage.filters.gaussian(ppp, 100))
NameError: name 'pp' is not defined
[14]:
m = img < skimage.filters.threshold_mean(img)
skimage.filters.threshold_mean((img * m).clip(np.min(img))).compute()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[14], line 1
----> 1 m = img < skimage.filters.threshold_mean(img)
2 skimage.filters.threshold_mean((img * m).clip(np.min(img))).compute()
NameError: name 'img' is not defined
[15]:
plt.imshow(img < skimage.filters.threshold_triangle(img))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[15], line 1
----> 1 plt.imshow(img < skimage.filters.threshold_triangle(img))
NameError: name 'img' is not defined
[16]:
def dabg(di):
m = di < skimage.filters.threshold_mean(di)
m1 = di < skimage.filters.threshold_mean((di * m).clip(np.min(di)))
# m2 = ndmorph.binary_dilation(~m1)
ndmorph.binary_dilation(~m1)
return da.ma.masked_array(di, mask=~m1)
def bg(im):
m = im < skimage.filters.threshold_mean(im)
m1 = im < skimage.filters.threshold_mean((im * m).clip(np.min(im)))
m2 = skimage.morphology.binary_dilation(~m1, footprint=np.ones([1, 1]))
# m2 = im < skimage.filters.threshold_triangle(np.ma.masked_array(im, mask=~m))
return np.ma.masked_array(im, mask=m2)
[17]:
tff.imshow(dabg(img).compute())
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[17], line 1
----> 1 tff.imshow(dabg(img).compute())
NameError: name 'img' is not defined
[18]:
flat = np.ma.mean(dabg(dd[333:500:1, 0]).compute(), axis=0)
skimage.io.imshow(flat)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[18], line 1
----> 1 flat = np.ma.mean(dabg(dd[333:500:1, 0]).compute(), axis=0)
2 skimage.io.imshow(flat)
NameError: name 'dd' is not defined
3.1.1. threshold mean clipping to min()#
[19]:
tff.imshow(bg(dd[0, 0]))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[19], line 1
----> 1 tff.imshow(bg(dd[0, 0]))
NameError: name 'dd' is not defined
[20]:
plt.hist(bg(img).ravel())
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[20], line 1
----> 1 plt.hist(bg(img).ravel())
NameError: name 'img' is not defined
[21]:
[np.ma.median(bg(dd[i, 0])) for i in range(10)]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[21], line 1
----> 1 [np.ma.median(bg(dd[i, 0])) for i in range(10)]
NameError: name 'dd' is not defined
[22]:
segmentation.bg_refine_iteratively(img.compute(), bgmax=40)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[22], line 1
----> 1 segmentation.bg_refine_iteratively(img.compute(), bgmax=40)
AttributeError: module 'nima.segmentation' has no attribute 'bg_refine_iteratively'
[23]:
%%time
np.ma.mean(bg(img.compute()))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
File <timed eval>:1
NameError: name 'img' is not defined
[24]:
%%time
utils.bg2(img.compute(), bgmax=60)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
File <timed eval>:1
AttributeError: module 'nima.utils' has no attribute 'bg2'
3.1.2. masked array (ma)#
[25]:
a = np.ma.masked_array([1, 4, 3], mask=[False, False, True])
b = np.ma.masked_array([10, 2, 6], mask=[False, True, False])
np.ma.median([a, b], axis=0)
[25]:
masked_array(data=[5.5, 3. , 4.5],
mask=False,
fill_value=1e+20)
[26]:
np.ma.median(np.ma.stack([a, b]), axis=0)
[26]:
masked_array(data=[5.5, 4.0, 6.0],
mask=[False, False, False],
fill_value=1e+20)
[27]:
f3 = img > skimage.filters.threshold_local(img.compute(), 601)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[27], line 1
----> 1 f3 = img > skimage.filters.threshold_local(img.compute(), 601)
NameError: name 'img' is not defined
[28]:
f3
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[28], line 1
----> 1 f3
NameError: name 'f3' is not defined
[29]:
img[~f3].mean().compute()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[29], line 1
----> 1 img[~f3].mean().compute()
NameError: name 'img' is not defined
[30]:
m1 = np.ma.masked_greater(img, 15)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[30], line 1
----> 1 m1 = np.ma.masked_greater(img, 15)
NameError: name 'img' is not defined
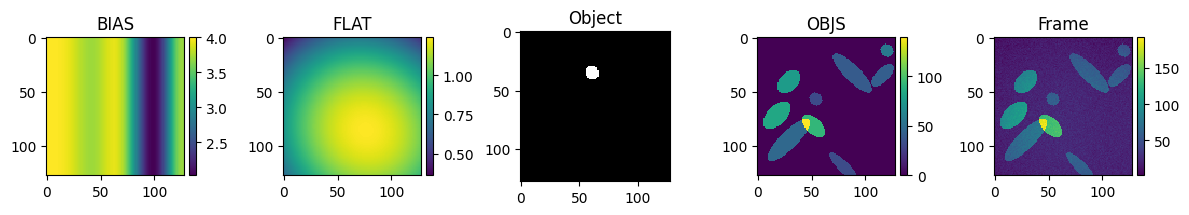
3.2. generat#
[31]:
image = "bias + noise + dark + flat * (sky + obj)"
image
[31]:
'bias + noise + dark + flat * (sky + obj)'
bias: generate a wave-like shape along x.
noise: random number will do.
dark: simply a scalar value.
flat: generate some 2D parabolic shape.
obj: circles-ellipsis. (MAYBE: like finite fractals to compare segmentation).
sky: None | some blurred circle-ellipsoid coincident and not with some obj.
fg_prj :=
bg_prj :=
[32]:
X = Y = 128
plt.figure(figsize=(12, 2.8))
plt.subplot(1, 5, 1)
plt.title("BIAS")
bias = generat.gen_bias(Y, X)
skimage.io.imshow(bias)
plt.subplot(1, 5, 2)
plt.title("FLAT")
flat = generat.gen_flat(Y, X)
skimage.io.imshow(flat)
plt.subplot(1, 5, 3)
plt.title("Object")
single_obj = generat.gen_object(Y, X, max_radius=7)
skimage.io.imshow(single_obj)
plt.subplot(1, 5, 4)
plt.title("OBJS")
objects = generat.gen_objs(
generat.ImageObjsParams(
max_fluor=100, max_num_objects=13, max_radius=12, min_radius=6, ncols=Y, nrows=X
)
)
skimage.io.imshow(objects)
plt.subplot(1, 5, 5)
plt.title("Frame")
frame = generat.gen_frame(objects, bias=bias, flat=flat, sky=17, noise_sd=3)
skimage.io.imshow(frame)
/tmp/ipykernel_2378/2707219899.py:8: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(bias)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/io/_plugins/matplotlib_plugin.py:158: UserWarning: Float image out of standard range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
/tmp/ipykernel_2378/2707219899.py:13: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(flat)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/io/_plugins/matplotlib_plugin.py:158: UserWarning: Float image out of standard range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
/tmp/ipykernel_2378/2707219899.py:18: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(single_obj)
/tmp/ipykernel_2378/2707219899.py:27: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(objects)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/io/_plugins/matplotlib_plugin.py:158: UserWarning: Float image out of standard range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
/tmp/ipykernel_2378/2707219899.py:32: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(frame)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/io/_plugins/matplotlib_plugin.py:158: UserWarning: Low image data range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
[32]:
<matplotlib.image.AxesImage at 0x7fade0fcf610>
[33]:
objects = generat.gen_objs(
generat.ImageObjsParams(
max_fluor=100,
max_num_objects=13,
max_radius=12,
min_radius=6,
ncols=Y,
nrows=X,
)
)
frame = generat.gen_frame(objects, bias=np.ones((Y, X)), flat=None, sky=7, noise_sd=6)
segmentation.bg_refine_iteratively(frame, probplot=1)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[33], line 12
1 objects = generat.gen_objs(
2 generat.ImageObjsParams(
3 max_fluor=100,
(...) 9 )
10 )
11 frame = generat.gen_frame(objects, bias=np.ones((Y, X)), flat=None, sky=7, noise_sd=6)
---> 12 segmentation.bg_refine_iteratively(frame, probplot=1)
AttributeError: module 'nima.segmentation' has no attribute 'bg_refine_iteratively'
[34]:
# Method {'arcsinh', 'entropy', 'adaptive', 'li_adaptive', 'li_li'} used for the
bg_result = segmentation.bg(frame, bg_params=segmentation.BgParams(kind="adaptive"))
bg_result.bg
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[34], line 2
1 # Method {'arcsinh', 'entropy', 'adaptive', 'li_adaptive', 'li_li'} used for the
----> 2 bg_result = segmentation.bg(frame, bg_params=segmentation.BgParams(kind="adaptive"))
3 bg_result.bg
AttributeError: module 'nima.segmentation' has no attribute 'bg'
[35]:
segmentation.bg_refine_iteratively(
frame, bgmax=segmentation._bgmax(frame, densityplot=True), probplot=True
)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[35], line 1
----> 1 segmentation.bg_refine_iteratively(
2 frame, bgmax=segmentation._bgmax(frame, densityplot=True), probplot=True
3 )
AttributeError: module 'nima.segmentation' has no attribute 'bg_refine_iteratively'
[36]:
r2 = utils.bg2(frame, bgmax=30)
plt.plot(r2[3])
r2[:2]
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[36], line 1
----> 1 r2 = utils.bg2(frame, bgmax=30)
3 plt.plot(r2[3])
4 r2[:2]
AttributeError: module 'nima.utils' has no attribute 'bg2'
[37]:
lim = np.arcsinh(frame)
plt.imshow(lim)
plt.colorbar()
[37]:
<matplotlib.colorbar.Colorbar at 0x7fadddf5c7d0>
[38]:
lim = ndimage.percentile_filter(lim, 80, size=10)
plt.imshow(lim)
plt.colorbar()
[38]:
<matplotlib.colorbar.Colorbar at 0x7faddddd9d10>
[39]:
bg_result = segmentation.bg_refine_iteratively(frame)
m0 = frame < bg_result.bg + 1 * bg_result.sd
plt.imshow(m0)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[39], line 1
----> 1 bg_result = segmentation.bg_refine_iteratively(frame)
2 m0 = frame < bg_result.bg + 1 * bg_result.sd
3 plt.imshow(m0)
AttributeError: module 'nima.segmentation' has no attribute 'bg_refine_iteratively'
[40]:
p = segmentation.prob(frame, bg_result.bg, bg_result.sd)
plt.imshow(p)
plt.colorbar()
plt.figure()
plt.plot(p[100, :], "o")
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[40], line 1
----> 1 p = segmentation.prob(frame, bg_result.bg, bg_result.sd)
2 plt.imshow(p)
3 plt.colorbar()
NameError: name 'bg_result' is not defined
[41]:
arcsinh_perc = 80
radius = 10
perc = 0.1
lim = np.arcsinh(frame)
lim = ndimage.percentile_filter(lim, arcsinh_perc, size=radius)
thr = (1 - perc) * lim.min() + perc * lim.max()
m = lim < thr
bgv = frame[m]
[42]:
plt.subplot(131)
plt.hist(p.ravel(), bins=99)
# plt.semilogy()
plt.subplot(132)
skimage.io.imshow(p)
plt.subplot(133)
skimage.io.imshow(p > 0.001)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[42], line 2
1 plt.subplot(131)
----> 2 plt.hist(p.ravel(), bins=99)
3 # plt.semilogy()
4 plt.subplot(132)
NameError: name 'p' is not defined
[43]:
prob_frame = segmentation.geometric_mean_filter(p, 1.9)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[43], line 1
----> 1 prob_frame = segmentation.geometric_mean_filter(p, 1.9)
NameError: name 'p' is not defined
[44]:
mask = prob_frame > 0.0029
segmentation.fit_gaussian(frame[mask]), scipy.stats.distributions.norm.fit(frame[mask])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[44], line 1
----> 1 mask = prob_frame > 0.0029
2 segmentation.fit_gaussian(frame[mask]), scipy.stats.distributions.norm.fit(frame[mask])
NameError: name 'prob_frame' is not defined
[45]:
bg, sd = bg_result.bg, bg_result.sd
mgeo = segmentation.geometric_mean_filter(utils.prob(frame, bg, sd), 1) > 0.01
# mgeo = skimage.filters.median(utils.prob(frame, bg, sd)) > 0.01
mgeo = (
ndimage.percentile_filter(utils.prob(frame, bg, sd), percentile=1, size=2) > 0.005
)
# mgeo = ndimage.uniform_filter(utils.prob(frame, bg222, sd222), size=1) > 0.005
# mgeo = ndimage.gaussian_filter(utils.prob(frame, bg222, sd222), 0.25) > 0.005
skimage.io.imshow(mgeo)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[45], line 1
----> 1 bg, sd = bg_result.bg, bg_result.sd
2 mgeo = segmentation.geometric_mean_filter(utils.prob(frame, bg, sd), 1) > 0.01
3 # mgeo = skimage.filters.median(utils.prob(frame, bg, sd)) > 0.01
NameError: name 'bg_result' is not defined
[46]:
objs = generat.gen_objs(
generat.ImageObjsParams(
max_fluor=15,
max_num_objects=20,
max_radius=12,
min_radius=6,
ncols=264,
nrows=64,
)
)
frame = generat.gen_frame(objs, None, None, sky=10, noise_sd=6)
bgres = nima.bg(frame.astype("float"))
# plt.hist(bgs, bins=8)
bgres.bg
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[46], line 13
1 objs = generat.gen_objs(
2 generat.ImageObjsParams(
3 max_fluor=15,
(...) 9 )
10 )
11 frame = generat.gen_frame(objs, None, None, sky=10, noise_sd=6)
---> 13 bgres = nima.bg(frame.astype("float"))
14 # plt.hist(bgs, bins=8)
15 bgres.bg
AttributeError: module 'nima.nima' has no attribute 'bg'
[47]:
segmentation.bg_refine_iteratively(frame, probplot=1)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[47], line 1
----> 1 segmentation.bg_refine_iteratively(frame, probplot=1)
AttributeError: module 'nima.segmentation' has no attribute 'bg_refine_iteratively'
simulation for:
varying error
varying number of cells
varying intensity of cells
in the absence of bias correction
in the absence of flat correction
[48]:
objs_pars = generat.ImageObjsParams(max_num_objects=40, max_fluor=15, nrows=64)
objs_pars
[48]:
ImageObjsParams(max_num_objects=40, nrows=64, ncols=128, min_radius=6, max_radius=12, max_fluor=15)
[49]:
utils.bg2(frame)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[49], line 1
----> 1 utils.bg2(frame)
AttributeError: module 'nima.utils' has no attribute 'bg2'
[50]:
bg_arcsinh = []
bg_entropy = []
bg_adaptive = []
bg_liadaptive = []
bg_lili = []
bg_invyen = []
bg_ = []
bg2_ = []
def safe_call(func, *args, **kwargs): # noqa: ANN002,ANN003
"""Wrap a function.
This wrapper attempts to call the given function with the provided arguments.
If the function call raises an exception, it returns NaN.
"""
try:
return func(*args, **kwargs)
except ZeroDivisionError:
print("Division by zero encountered.")
return np.nan
except Exception as e: # noqa: BLE001
print(f"An error occurred: {e}")
return np.nan
objs_pars = generat.ImageObjsParams(max_num_objects=10, max_fluor=100, nrows=128)
noise_sd = 4
for _ in range(1):
print(_)
objs = generat.gen_objs(objs_pars)
frame = generat.gen_frame(objs, None, None, sky=10, noise_sd=noise_sd)
bg_arcsinh.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="arcsinh")).bg
)
bg_entropy.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="entropy")).bg
)
bg_adaptive.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="adaptive")).bg
)
bg_liadaptive.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="li_adaptive")).bg
)
bg_lili.append(safe_call(nima.bg, frame, segmentation.BgParams(kind="li_li")).bg)
# bg_invyen.append(safe_call(nima.bg, frame, segmentation.BgParams(kind="inverse_yen")).bg)
bg_.append(safe_call(segmentation.bg_refine_iteratively, frame).bg)
bg2_.append(safe_call(utils.bg2, frame, bgmax=500)[0])
0
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[50], line 35
32 objs = generat.gen_objs(objs_pars)
33 frame = generat.gen_frame(objs, None, None, sky=10, noise_sd=noise_sd)
34 bg_arcsinh.append(
---> 35 safe_call(nima.bg, frame, segmentation.BgParams(kind="arcsinh")).bg
36 )
37 bg_entropy.append(
38 safe_call(nima.bg, frame, segmentation.BgParams(kind="entropy")).bg
39 )
40 bg_adaptive.append(
41 safe_call(nima.bg, frame, segmentation.BgParams(kind="adaptive")).bg
42 )
AttributeError: module 'nima.nima' has no attribute 'bg'
[51]:
def sim2df(num_of_repeats, noise_sd, objs_pars):
bg_arcsinh = []
bg_entropy = []
bg_adaptive = []
bg_liadaptive = []
bg_lili = []
bg_invyen = []
bg_ = []
bg2_ = []
for _ in range(num_of_repeats):
# print(_)
objs = generat.gen_objs(objs_pars)
frame = generat.gen_frame(objs, None, None, sky=10, noise_sd=noise_sd)
bg_arcsinh.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="arcsinh")).bg
)
bg_entropy.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="entropy")).bg
)
bg_adaptive.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="adaptive")).bg
)
bg_liadaptive.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="li_adaptive")).bg
)
bg_lili.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="li_li")).bg
)
bg_invyen.append(
safe_call(nima.bg, frame, segmentation.BgParams(kind="inverse_yen")).bg
)
bg_.append(safe_call(segmentation.bg_refine_iteratively, frame).bg)
bg2_.append(safe_call(utils.bg2, frame, bgmax=800)[0])
df = pd.DataFrame(
np.column_stack(
(
bg_arcsinh,
bg_entropy,
bg_adaptive,
bg_liadaptive,
bg_lili,
bg_invyen,
bg_,
bg2_,
)
),
columns=[
"arcsinh",
"entropy",
"adaptive",
"li_adaptive",
"li li",
"inv_yen",
"bg",
"bg2",
],
)
df["sd"] = noise_sd
return df
[52]:
objs_pars = generat.ImageObjsParams(max_num_objects=10, max_fluor=100, nrows=128)
# df_all
[53]:
df_all = pd.DataFrame()
for noise_sd in [1, 2, 3, 4]:
df = sim2df(num_of_repeats=49, objs_pars=objs_pars, noise_sd=noise_sd)
df_all = pd.concat([df_all, df])
f = df_all.boxplot(vert=False, showfliers=0, by="sd")
plt.xlim((5, 15))
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[53], line 3
1 df_all = pd.DataFrame()
2 for noise_sd in [1, 2, 3, 4]:
----> 3 df = sim2df(num_of_repeats=49, objs_pars=objs_pars, noise_sd=noise_sd)
4 df_all = pd.concat([df_all, df])
6 f = df_all.boxplot(vert=False, showfliers=0, by="sd")
Cell In[51], line 16, in sim2df(num_of_repeats, noise_sd, objs_pars)
13 objs = generat.gen_objs(objs_pars)
14 frame = generat.gen_frame(objs, None, None, sky=10, noise_sd=noise_sd)
15 bg_arcsinh.append(
---> 16 safe_call(nima.bg, frame, segmentation.BgParams(kind="arcsinh")).bg
17 )
18 bg_entropy.append(
19 safe_call(nima.bg, frame, segmentation.BgParams(kind="entropy")).bg
20 )
21 bg_adaptive.append(
22 safe_call(nima.bg, frame, segmentation.BgParams(kind="adaptive")).bg
23 )
AttributeError: module 'nima.nima' has no attribute 'bg'
[54]:
import seaborn as sns
[55]:
dfmelted = df_all.melt(ignore_index=1, id_vars="sd")
dfmelted.head()
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[55], line 1
----> 1 dfmelted = df_all.melt(ignore_index=1, id_vars="sd")
2 dfmelted.head()
File ~/work/nima/nima/.venv/lib/python3.13/site-packages/pandas/core/frame.py:9969, in DataFrame.melt(self, id_vars, value_vars, var_name, value_name, col_level, ignore_index)
9959 @Appender(_shared_docs["melt"] % {"caller": "df.melt(", "other": "melt"})
9960 def melt(
9961 self,
(...) 9967 ignore_index: bool = True,
9968 ) -> DataFrame:
-> 9969 return melt(
9970 self,
9971 id_vars=id_vars,
9972 value_vars=value_vars,
9973 var_name=var_name,
9974 value_name=value_name,
9975 col_level=col_level,
9976 ignore_index=ignore_index,
9977 ).__finalize__(self, method="melt")
File ~/work/nima/nima/.venv/lib/python3.13/site-packages/pandas/core/reshape/melt.py:74, in melt(frame, id_vars, value_vars, var_name, value_name, col_level, ignore_index)
70 if missing.any():
71 missing_labels = [
72 lab for lab, not_found in zip(labels, missing) if not_found
73 ]
---> 74 raise KeyError(
75 "The following id_vars or value_vars are not present in "
76 f"the DataFrame: {missing_labels}"
77 )
78 if value_vars_was_not_none:
79 frame = frame.iloc[:, algos.unique(idx)]
KeyError: "The following id_vars or value_vars are not present in the DataFrame: ['sd']"
[ ]:
[56]:
sns.barplot(dfmelted, x="variable", y="value", hue="sd", dodge=1, palette="Blues_d")
plt.ylim(4, 10.5)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[56], line 1
----> 1 sns.barplot(dfmelted, x="variable", y="value", hue="sd", dodge=1, palette="Blues_d")
2 plt.ylim(4, 10.5)
NameError: name 'dfmelted' is not defined
[57]:
sns.stripplot(
dfmelted,
x="variable",
y="value",
hue="sd",
dodge=1,
jitter=0.2,
alpha=0.6,
palette="Reds_d",
)
sns.boxenplot(
dfmelted, x="variable", y="value", hue="sd", dodge=1, alpha=0.6, palette="Reds_d"
)
plt.ylim(7.0, 11)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[57], line 2
1 sns.stripplot(
----> 2 dfmelted,
3 x="variable",
4 y="value",
5 hue="sd",
6 dodge=1,
7 jitter=0.2,
8 alpha=0.6,
9 palette="Reds_d",
10 )
11 sns.boxenplot(
12 dfmelted, x="variable", y="value", hue="sd", dodge=1, alpha=0.6, palette="Reds_d"
13 )
15 plt.ylim(7.0, 11)
NameError: name 'dfmelted' is not defined
[58]:
skimage.io.imshow(frame)
/tmp/ipykernel_2378/618435722.py:1: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(frame)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/io/_plugins/matplotlib_plugin.py:158: UserWarning: Low image data range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
[58]:
<matplotlib.image.AxesImage at 0x7faddcac7610>
[59]:
import seaborn as sns
sns.regplot(pd.DataFrame({"x": bg_arcsinh, "y": bg_}), x="x", y="y")
[59]:
<Axes: xlabel='x', ylabel='y'>
[60]:
# Generate a single small object
small_object = generat.gen_object(nrows=12, ncols=12, min_radius=3, max_radius=4)
# Plot the generated object
plt.imshow(small_object, cmap="gray")
plt.title("Single Small Object")
plt.xlabel("X")
plt.ylabel("Y")
plt.show()
[61]:
import scipy.signal
# Convolve the small object with the flat image
convolved_image = scipy.signal.convolve2d(flat, small_object, mode="same")
# Plot the convolved image
plt.imshow(convolved_image, cmap="gray")
plt.colorbar()
plt.title("Convolved Image")
[61]:
Text(0.5, 1.0, 'Convolved Image')
[62]:
flat.shape[1] - small_object.shape[1]
[62]:
116
[63]:
# Number of frames in the stack
num_frames = 10000
# Initialize an empty stack to store the frames
stack = np.zeros_like(flat)
# Iterate over each frame in the stack
for _ in range(num_frames):
# Generate random coordinates for the position of the small object within the flat image
x_pos = nrg.integers(0, flat.shape[1] - small_object.shape[1])
y_pos = nrg.integers(0, flat.shape[0] - small_object.shape[0])
# Add the small object to the flat image at the random position
flat_image_with_object = flat.copy()
flat_image_with_object[
y_pos : y_pos + small_object.shape[0], x_pos : x_pos + small_object.shape[1]
] += small_object
# Add the frame with the small object to the stack
stack += flat_image_with_object
# Plot the summed stack
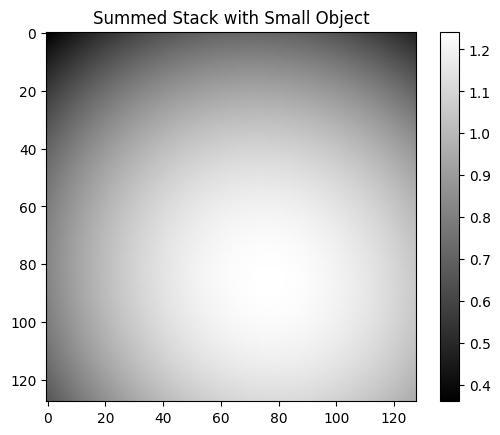
estimated = stack / stack.mean()
plt.imshow(estimated, cmap="gray")
plt.colorbar()
plt.title("Summed Stack with Small Object")
plt.show()
# plt.imshow(estimated - flat, cmap='gray')
skimage.io.imshow(ndimage.gaussian_filter(estimated, sigma=3) - flat)
/tmp/ipykernel_2378/2446578482.py:29: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(ndimage.gaussian_filter(estimated, sigma=3) - flat)
[63]:
<matplotlib.image.AxesImage at 0x7faddca2b390>
[64]:
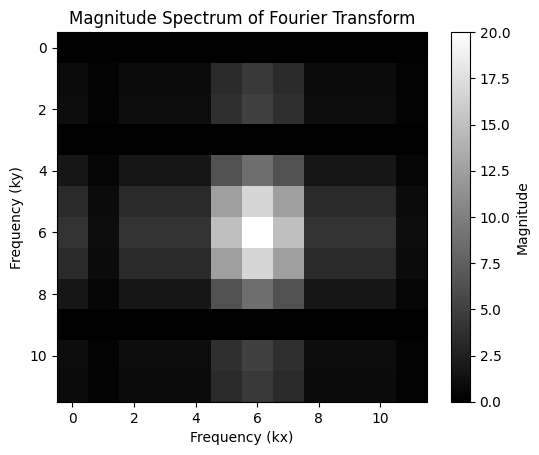
# Calculate the Fourier transform of the small object
fourier_transform_obj = np.fft.fft2(small_object)
# Calculate the magnitude spectrum of the Fourier transform
magnitude_spectrum = np.abs(np.fft.fftshift(fourier_transform_obj))
# Plot the magnitude spectrum
plt.imshow(magnitude_spectrum, cmap="gray")
plt.colorbar(label="Magnitude")
plt.title("Magnitude Spectrum of Fourier Transform")
plt.xlabel("Frequency (kx)")
plt.ylabel("Frequency (ky)")
plt.show()
[65]:
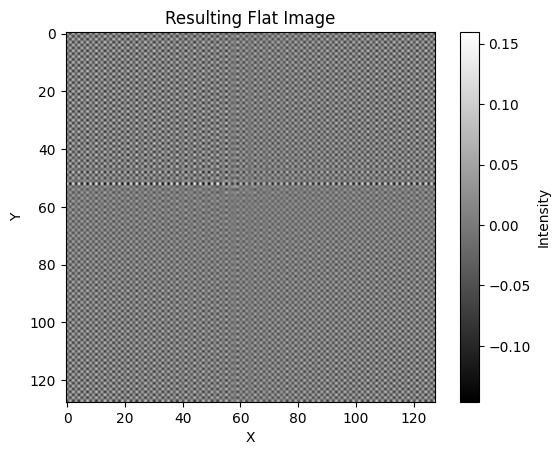
# Apply the convolution theorem
flat_fft = np.fft.fft2(stack)
# Calculate the Fourier transform of the small object
fourier_transform_obj = np.fft.fft2(small_object)
# Pad the small object to match the shape of flat
padded_obj = np.pad(
small_object,
(
(0, flat.shape[0] - small_object.shape[0]),
(0, flat.shape[1] - small_object.shape[1]),
),
mode="constant",
)
# Calculate the Fourier transform of the padded small object
fourier_transform_padded_obj = np.fft.fft2(padded_obj)
# Calculate the Fourier transform of the flat image
flat_fft = np.fft.fft2(flat)
# Perform element-wise division
result_fft = np.fft.ifftshift(
np.fft.ifft2(np.fft.fftshift(flat_fft / fourier_transform_padded_obj))
)
# result_fft = np.fft.ifftshift(np.fft.ifft2(np.fft.fftshift(flat_fft / fourier_transform_obj)))
# Take the real part to get rid of any numerical artifacts
result = np.real(result_fft)
# Plot the resulting flat image
plt.imshow(result, cmap="gray")
plt.colorbar(label="Intensity")
plt.title("Resulting Flat Image")
plt.xlabel("X")
plt.ylabel("Y")
plt.show()
[66]:
from nima import generat
flat = generat.gen_flat()
bias = 7 * np.ones((128, 128))
objs = generat.gen_objs(generat.ImageObjsParams(max_fluor=20, max_num_objects=80))
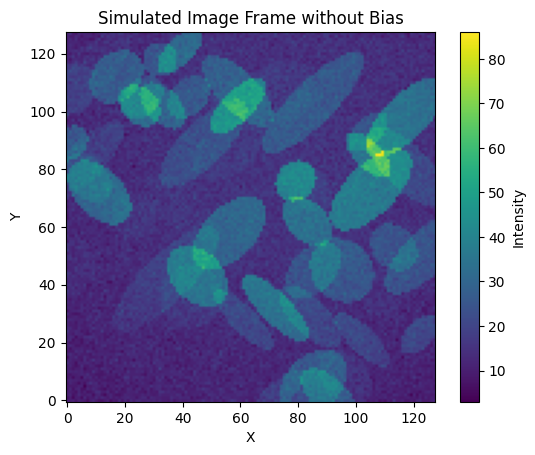
frame = generat.gen_frame(objs, bias=bias, flat=flat, noise_sd=2, sky=7)
# Plot the frame
plt.imshow(frame, cmap="viridis", origin="lower")
plt.colorbar(label="Intensity")
plt.title("Simulated Image Frame without Bias")
plt.xlabel("X")
plt.ylabel("Y")
plt.show()
[67]:
from tqdm import tqdm
# Generate a stack of frames
num_frames = 100
frame_stack = []
for _ in tqdm(range(num_frames), desc="Generating Frames"):
objs = generat.gen_objs(generat.ImageObjsParams(max_fluor=20, max_num_objects=80))
frame = generat.gen_frame(objs, bias=bias, flat=flat, noise_sd=2, sky=7)
frame_stack.append(frame)
Generating Frames: 100%|██████████| 100/100 [00:00<00:00, 125.12it/s]
[68]:
from functools import partial
p999 = partial(np.percentile, q=99.7)
p999.__name__ = "percentile 99.9%"
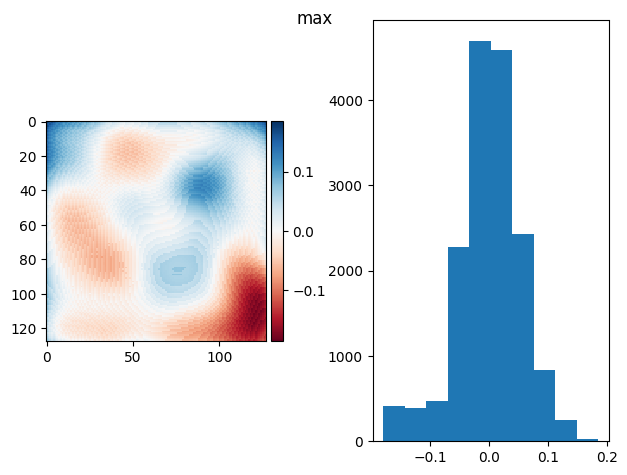
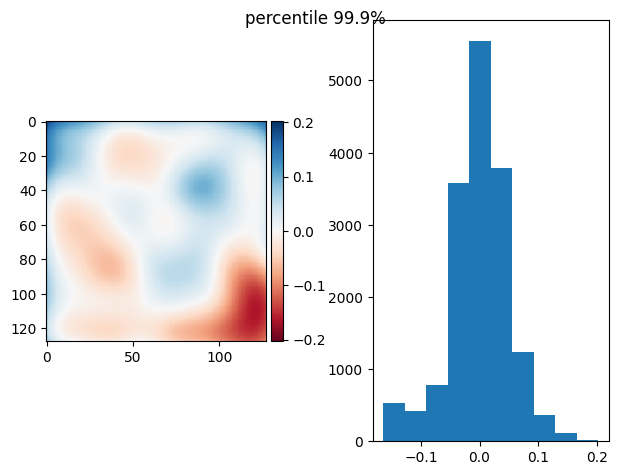
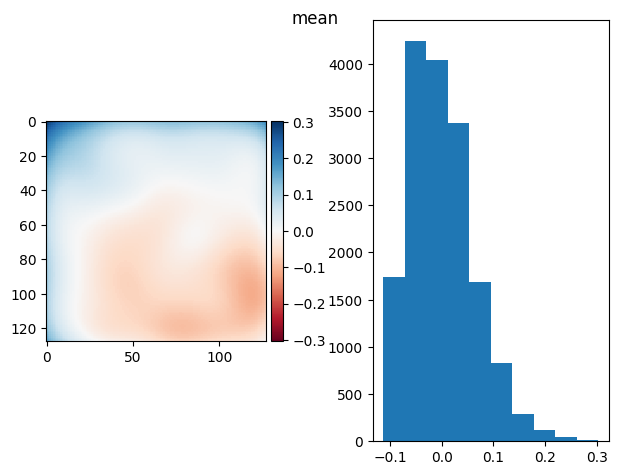
def diff_plot(im, flat, title):
f, axs = plt.subplots(1, 2)
diff = im / im.mean() - flat
skimage.io.imshow(diff, ax=axs[0])
axs[1].hist(diff.ravel())
f.suptitle(title)
return diff.mean(), diff.std()
def prj_plot(t_prj, title, sigma=128 / 11):
im = ndimage.gaussian_filter(t_prj, sigma=sigma)
return diff_plot(im, flat, title)
def prj(stack, func):
t_prj = func(stack, axis=0)
return prj_plot(t_prj, func.__name__)
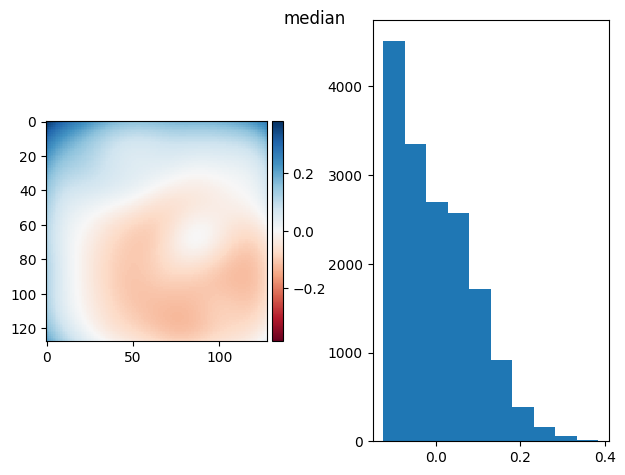
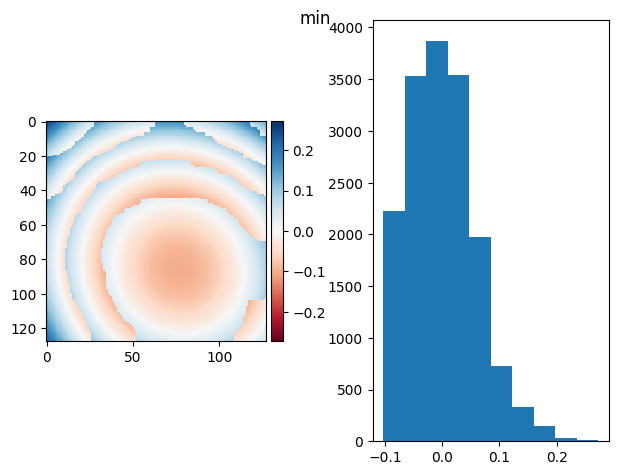
prj(frame_stack, np.max)
prj(frame_stack, p999)
prj(frame_stack, np.mean)
prj(frame_stack, np.median)
prj(frame_stack, np.min)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[68]:
(np.float64(1.0234868508263162e-16), np.float64(0.05749263872023587))
[69]:
objs = generat.gen_objs(generat.ImageObjsParams(max_fluor=20, max_num_objects=8))
frame = generat.gen_frame(objs, bias=bias, flat=flat)
plt.imshow(frame)
[69]:
<matplotlib.image.AxesImage at 0x7faddc113ed0>
[70]:
bias = np.zeros((128, 128))
flat = np.ones((128, 128))
stack = np.stack(
[
generat.gen_frame(
generat.gen_objs(generat.ImageObjsParams(max_fluor=20, max_num_objects=20)),
None,
None,
noise_sd=8,
sky=20,
)
for i in range(1000)
]
)
[71]:
stat_bg = []
stat_sd = []
for s in stack[:]:
bg, sd = segmentation.iteratively_refine_background(s)[:2]
stat_bg.append(bg)
stat_sd.append(sd)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[71], line 4
2 stat_sd = []
3 for s in stack[:]:
----> 4 bg, sd = segmentation.iteratively_refine_background(s)[:2]
5 stat_bg.append(bg)
6 stat_sd.append(sd)
AttributeError: module 'nima.segmentation' has no attribute 'iteratively_refine_background'
[72]:
plt.subplot(121)
plt.hist(stat_bg)
print(np.mean(stat_bg), np.std(stat_bg))
plt.subplot(122)
plt.hist(stat_sd)
np.mean(stat_sd), np.std(stat_sd)
nan nan
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/numpy/_core/fromnumeric.py:3860: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/numpy/_core/_methods.py:144: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/numpy/_core/_methods.py:222: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/numpy/_core/_methods.py:180: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/numpy/_core/_methods.py:214: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
[72]:
(np.float64(nan), np.float64(nan))
[73]:
plt.imshow(stack[1])
[73]:
<matplotlib.image.AxesImage at 0x7fadd4ef9090>
3.3. what is the best projection for flat calculation?#
[74]:
bias = generat.gen_bias()
flat = generat.gen_flat()
stack = np.stack(
[
generat.gen_frame(
generat.gen_objs(generat.ImageObjsParams(max_fluor=20)),
bias,
flat,
noise_sd=1,
sky=2,
)
for i in range(1000)
]
)
[75]:
def splot(stack, num=4):
_f, axs = plt.subplots(1, num)
for i in range(num):
axs[i].imshow(stack[nrg.integers(0, len(stack))])
splot(stack)
[76]:
prj(stack, np.max)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[76]:
(np.float64(6.678685382510707e-17), np.float64(0.07373137780750438))
[77]:
prj(stack, np.mean)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[77]:
(np.float64(7.28583859910259e-17), np.float64(0.15313720092367075))
[78]:
prj(stack, np.median)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[78]:
(np.float64(-9.367506770274758e-17), np.float64(0.17348169468639787))
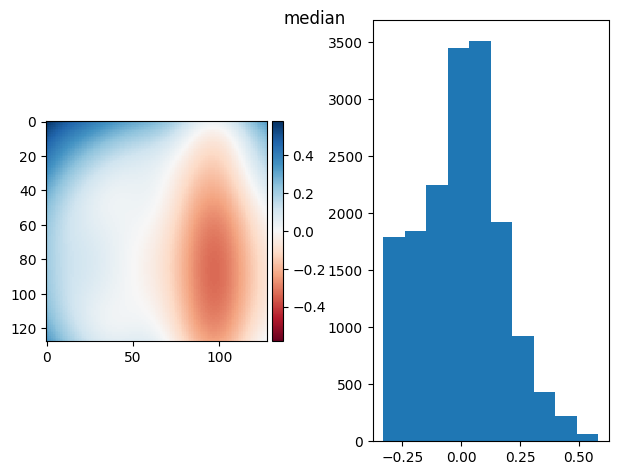
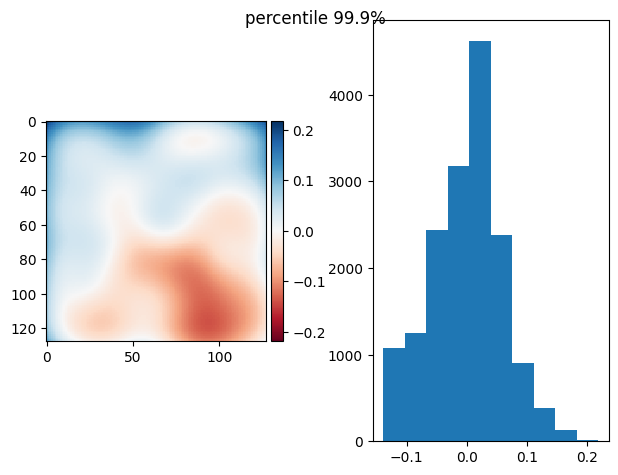
[79]:
from functools import partial
p999 = partial(np.percentile, q=99.9)
p999.__name__ = "percentile 99.9%"
prj(stack, p999)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[79]:
(np.float64(-1.214306433183765e-17), np.float64(0.06664066745904444))
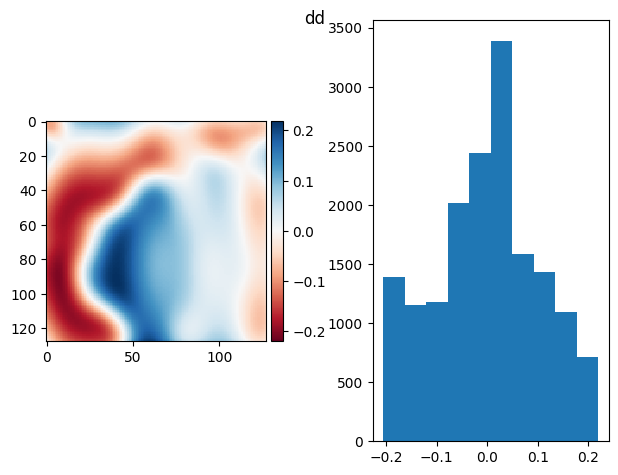
[80]:
im = np.mean(
ndfilters.median_filter(
da.from_array(stack[:100] - bias), size=(32, 16, 16)
).compute(),
axis=0,
)
prj_plot(im, "dd", sigma=7)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[80]:
(np.float64(5.898059818321144e-17), np.float64(0.10518590675429572))
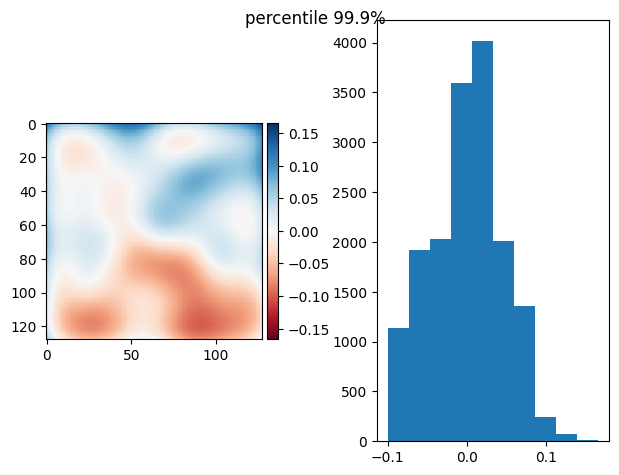
[81]:
prj(stack - bias, p999)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[81]:
(np.float64(9.020562075079397e-17), np.float64(0.045588248784387615))
[82]:
prj(stack - bias, np.mean)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[82]:
(np.float64(4.683753385137379e-17), np.float64(0.035219771633705675))
3.3.1. Using fg and bg masks?#
And assuming we know the bias of the camera.
[83]:
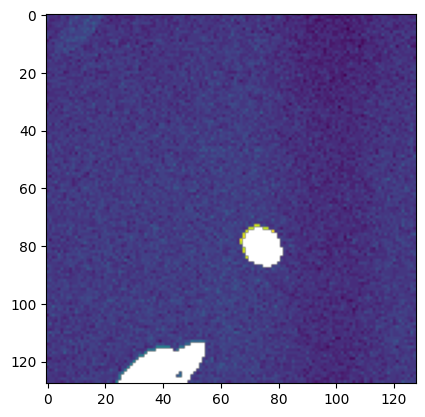
def mask_plane(plane, bg_ave=2, bg_std=1.19, erf_pvalue=0.01):
p = utils.prob(plane, bg_ave, bg_std)
p = ndimage.median_filter(p, size=2)
mask = p > erf_pvalue
mask = skimage.morphology.remove_small_holes(mask)
return np.ma.masked_array(plane, mask=~mask), np.ma.masked_array(plane, mask=mask)
plt.imshow(mask_plane(stack[113], *utils.bg(stack[113]))[0])
[83]:
<matplotlib.image.AxesImage at 0x7faddc0c91d0>
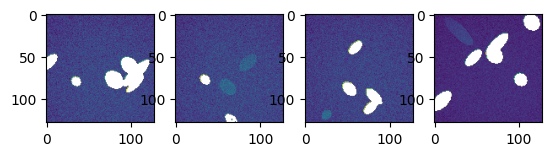
[84]:
bgs, fgs = list(
zip(*[mask_plane(s - bias, *utils.bg(s - bias)) for s in stack], strict=False)
)
splot(bgs)
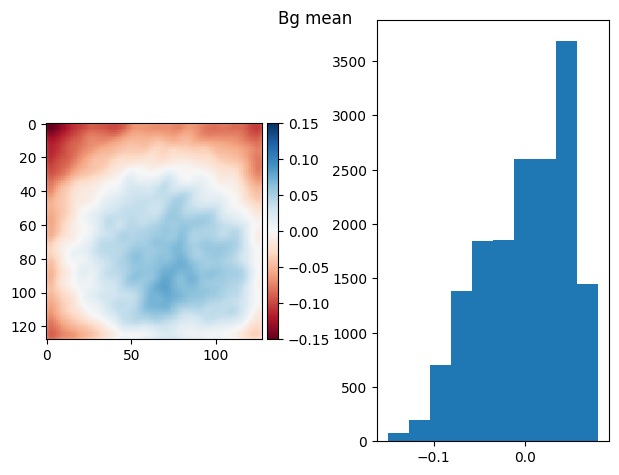
[85]:
t_prj = np.ma.mean(np.ma.stack(bgs), axis=0)
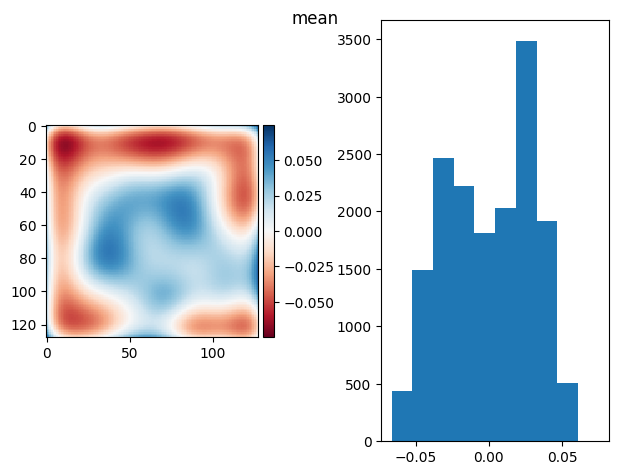
prj_plot(t_prj, "Bg mean", sigma=3)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[85]:
(np.float64(1.734723475976807e-16), np.float64(0.04835694053367755))
[86]:
t_prj = np.ma.max(np.ma.stack(fgs), axis=0)
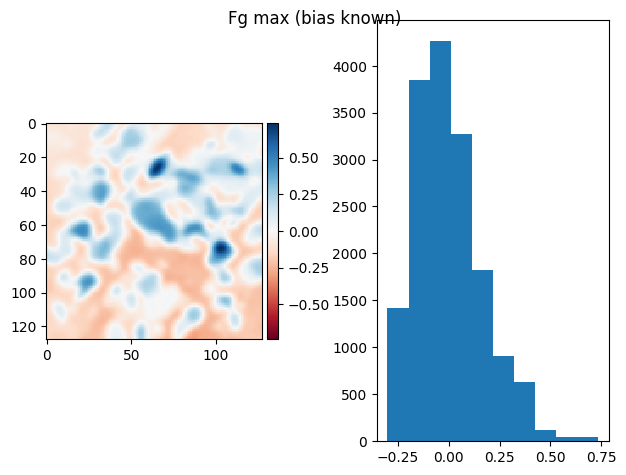
prj_plot(t_prj, "Fg max (bias known)", sigma=2)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[86]:
(np.float64(1.2663481374630692e-16), np.float64(0.15844019690040237))
[87]:
bgs, fgs = list(zip(*[mask_plane(s, *utils.bg(s)) for s in stack], strict=False))
bg_prj1 = np.ma.mean(np.ma.stack(bgs[:]), axis=0)
fg_prj1 = np.ma.mean(np.ma.stack(fgs[:]), axis=0)
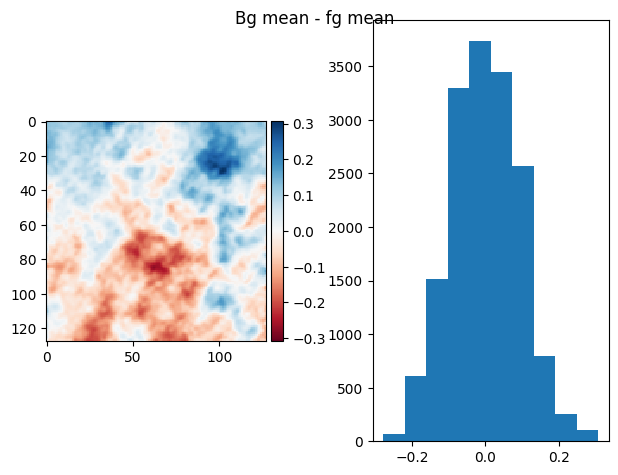
im = fg_prj1 - bg_prj1
diff_plot(ndimage.gaussian_filter(im, 1), flat, "Bg mean - fg mean")
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[87]:
(np.float64(6.245004513516506e-17), np.float64(0.07799793200955611))
[88]:
bg_prj = np.ma.mean(bgs, axis=0)
fg_prj = np.ma.max(fgs, axis=0)
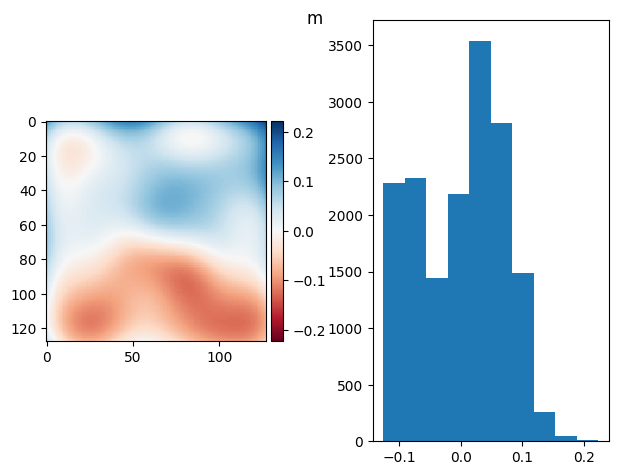
# im = ndimage.median_filter(bg_prj-fg_prj, size=60) #- 2 * flat
im = ndimage.gaussian_filter(bg_prj - fg_prj, sigma=14) # - 2 * flat
diff_plot(im, flat, "m")
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[88]:
(np.float64(3.642919299551295e-17), np.float64(0.06347681088430922))
[89]:
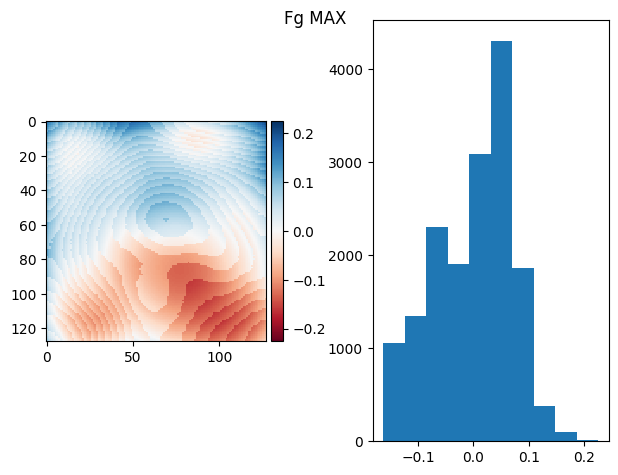
t_prj = np.ma.max(fgs, axis=0)
prj_plot(t_prj, "Fg MAX", sigma=13)
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[89]:
(np.float64(7.112366251504909e-17), np.float64(0.0737124310575951))
[90]:
eflat = bg_prj - fg_prj
eflat /= eflat.mean()
eflat = ndimage.gaussian_filter(eflat, sigma=13)
diff_plot(eflat, flat, "eflat")
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
[90]:
(np.float64(-2.5153490401663703e-17), np.float64(0.06420304793008824))
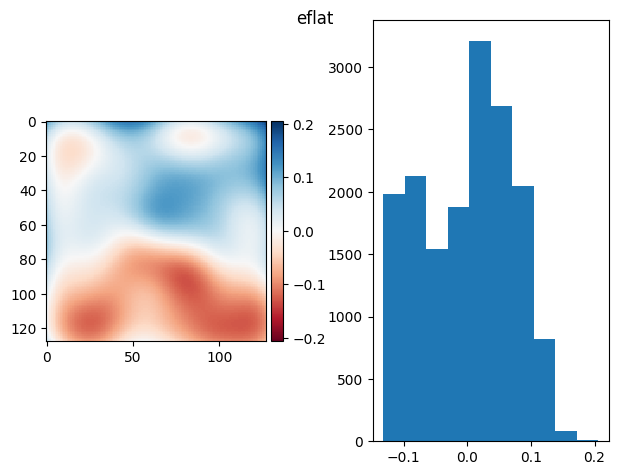
3.4. When bias and flat are unknown…#
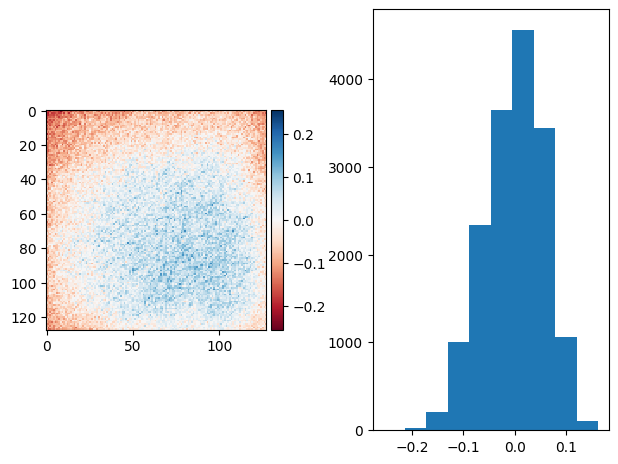
bias = bg_prj - sky * flat
bias = fg_prj - flat
sky * flat - flat = bg_prj - fg_prj
[91]:
diff_plot((bg_prj1 - bias) / 2, flat, "")
/tmp/ipykernel_2378/975262232.py:10: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(diff, ax=axs[0])
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/numpy/lib/_function_base_impl.py:4859: UserWarning: Warning: 'partition' will ignore the 'mask' of the MaskedArray.
arr.partition(
[91]:
(np.float64(1.0755285551056204e-16), np.float64(0.05794589230665674))
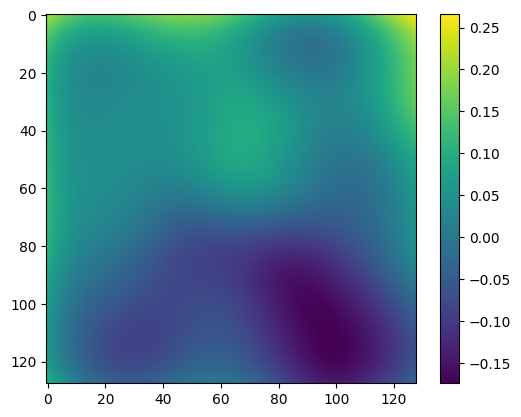
[92]:
plt.imshow((im - bias) / (im - bias).mean() - flat)
plt.colorbar()
[92]:
<matplotlib.colorbar.Colorbar at 0x7faddc3d6d50>
3.5. cfr. nima.bg#
[93]:
# r = nima.bg((stack[113] - bias) / flat)
r = nima.bg(stack[111])
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[93], line 2
1 # r = nima.bg((stack[113] - bias) / flat)
----> 2 r = nima.bg(stack[111])
AttributeError: module 'nima.nima' has no attribute 'bg'
[94]:
r[1].mean(), r[1].std()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[94], line 1
----> 1 r[1].mean(), r[1].std()
NameError: name 'r' is not defined
[95]:
utils.bg(stack[111])
[95]:
(np.float64(5.458047033968789), np.float64(1.2255365147174444))
[96]:
bias.mean() + 2
[96]:
np.float64(5.418070740426731)
3.6. geometric mean#
[97]:
vals = [0.8, 0.1, 0.3, 0.1, 0.8, 0.8, 0.8, 0.1, 0.8]
np.median(vals), scipy.stats.gmean(vals), np.mean(vals)
[97]:
(np.float64(0.8),
np.float64(0.35869897213131746),
np.float64(0.5111111111111111))
[98]:
p = vals[0]
p *= vals[1]
p *= vals[2]
p *= vals[3]
p *= vals[4]
p *= vals[5]
p *= vals[6]
p *= vals[7]
p *= vals[8]
np.power(p, 1 / 9)
[98]:
np.float64(0.3586989721313174)
[99]:
np.exp(np.log(np.array(vals)).sum() / 9)
[99]:
np.float64(0.35869897213131746)
[100]:
vv = np.array(vals).reshape(3, 3)
vv
[100]:
array([[0.8, 0.1, 0.3],
[0.1, 0.8, 0.8],
[0.8, 0.1, 0.8]])
[101]:
segmentation.geometric_mean_filter(vv, 1.0)
5.0
[101]:
array([[0.38073079, 0.45358663, 0.47428812],
[0.55189186, 0.22973967, 0.68750877],
[0.38073079, 0.55189186, 0.57707996]])
[102]:
kernel = skimage.morphology.disk(1.0).astype(float)
n = np.sum(kernel) # Total weight, or number of ones in the kernel
print(n)
plt.imshow(kernel)
5.0
[102]:
<matplotlib.image.AxesImage at 0x7faddc870910>
[103]:
(0.8 * 0.8 * 0.1 * 0.1 * 0.1) ** (1 / 5)
[103]:
0.229739670999407