1. Usage#
[1]:
%load_ext autoreload
%autoreload 2
import matplotlib.pyplot as plt
import numpy as np
import scipy
import seaborn as sns
import skimage
import skimage.io
import tifffile
from pympler.asizeof import asizeof
from nima import nima, segmentation
cm = plt.cm.inferno_r
fp = "../../tests/data/1b_c16_15.tif"
channels = ["G", "R", "C"]
# dark = io.imread('/home/dati/GBM_persson/analyses/15.02.05_cal-GBM5-pBJclop/dark/dark-25_500.tif')
# flat = io.imread('/home/dati/GBM_persson/analyses/15.02.05_cal-GBM5-pBJclop/flat/flat-C-dark-37bis_500.tif')
[2]:
bg_params = segmentation.BgParams(kind="li_adaptive")
bg_params
[2]:
BgParams(kind='li_adaptive', perc=0.1, radius=10, adaptive_radius=None, arcsinh_perc=80, erosion_disk=0, clip=False)
[3]:
d_im, n_ch, times = nima.read_tiff(fp, channels)
(4, 512, 512)
[4]:
d_im_bg, bgs, ff = nima.d_bg(d_im, downscale=(2, 2), bg_params=bg_params)
bgs
[4]:
| G | R | C | |
|---|---|---|---|
| 0 | 462.00 | 249.75 | 278.00 |
| 1 | 463.50 | 253.00 | 280.25 |
| 2 | 464.50 | 254.75 | 282.00 |
| 3 | 461.25 | 252.50 | 278.00 |
[5]:
d_im_bg["G"].__sizeof__() / 1024**2
[5]:
8.000137329101562
[6]:
ff["C"][1][0]
[6]:
[7]:
nima.d_mask_label(
d_im_bg, threshold_method="yen", min_size=2000, channels=channels, watershed=0
)
d_im_bg.keys()
[7]:
dict_keys(['G', 'R', 'C', 'mask', 'labels'])
[8]:
f = nima.d_show(d_im_bg)
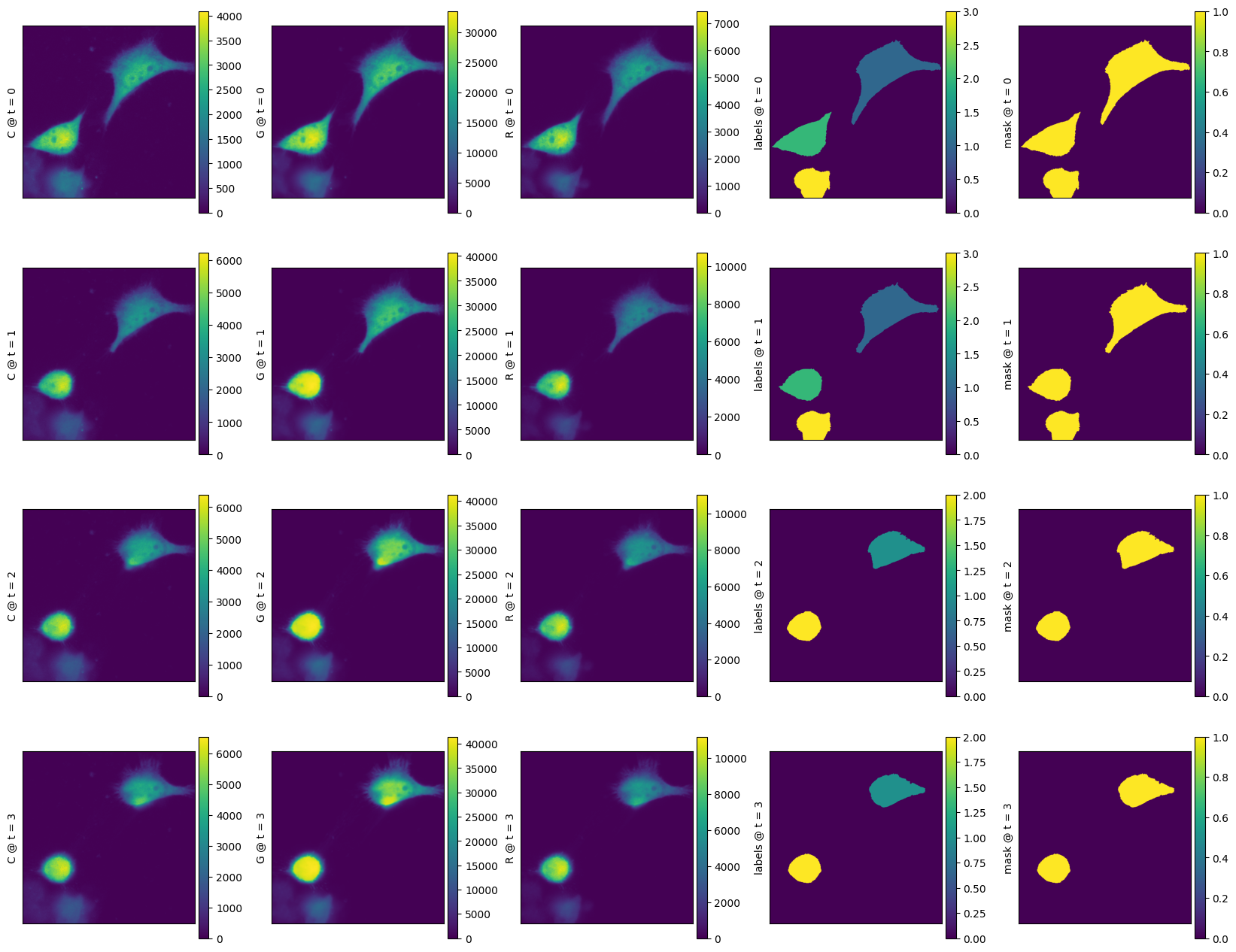
[9]:
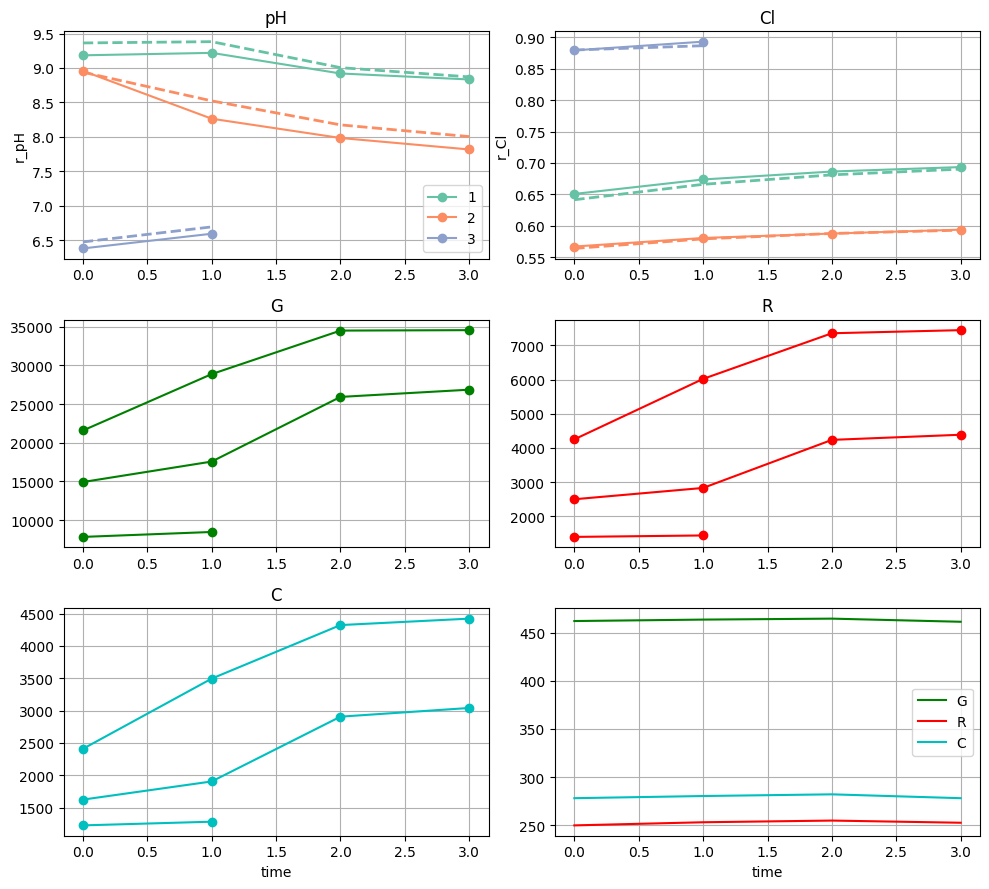
meas, pr = nima.d_meas_props(d_im_bg)
f = nima.d_plot_meas(bgs, meas, channels=channels)
[10]:
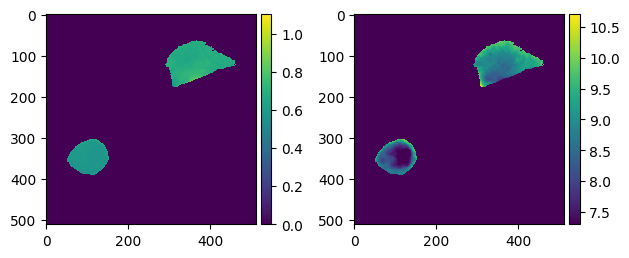
plt.subplot(1, 2, 1)
skimage.io.imshow(d_im_bg["r_cl"][2], vmin=0.0, vmax=1.1)
plt.subplot(1, 2, 2)
skimage.io.imshow(d_im_bg["r_pH"][2], vmin=7.3, vmax=10.7)
/tmp/ipykernel_2665/3384179216.py:2: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(d_im_bg["r_cl"][2], vmin=0.0, vmax=1.1)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/io/_plugins/matplotlib_plugin.py:158: UserWarning: Float image out of standard range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
/tmp/ipykernel_2665/3384179216.py:4: FutureWarning: `imshow` is deprecated since version 0.25 and will be removed in version 0.27. Please use `matplotlib`, `napari`, etc. to visualize images.
skimage.io.imshow(d_im_bg["r_pH"][2], vmin=7.3, vmax=10.7)
/home/runner/work/nima/nima/.venv/lib/python3.13/site-packages/skimage/io/_plugins/matplotlib_plugin.py:158: UserWarning: Float image out of standard range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
[10]:
<matplotlib.image.AxesImage at 0x7f5af8f66350>
1.1. dev#
[11]:
c0 = d_im["C"][0]
g0 = d_im["G"][0]
r0 = d_im["R"][0]
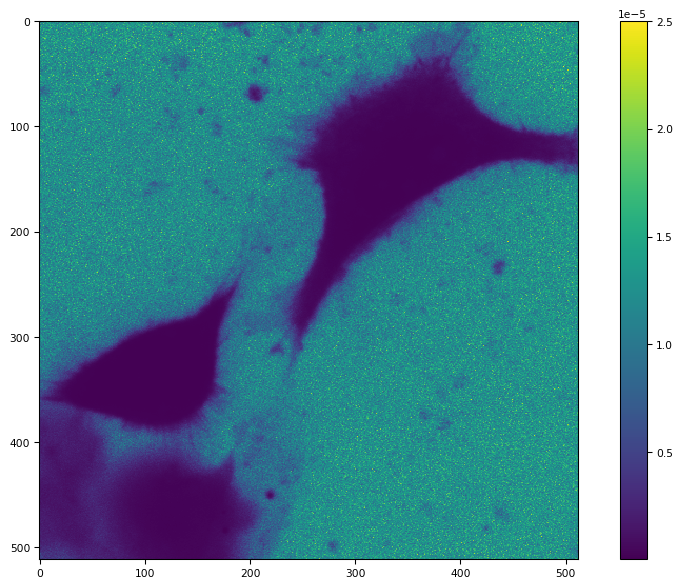
tifffile.imshow(1 / c0 / c0)
[11]:
(<Figure size 988.8x604.8 with 2 Axes>,
<Axes: >,
<matplotlib.image.AxesImage at 0x7f5af8e26210>)
[12]:
bg_params = segmentation.BgParams(kind="li_adaptive", erosion_disk=0)
bg_params
[12]:
BgParams(kind='li_adaptive', perc=0.1, radius=10, adaptive_radius=None, arcsinh_perc=80, erosion_disk=0, clip=False)
[13]:
im = c0
im = d_im["C"][1]
bg_params.adaptive_radius = int(im.shape[1] / 2)
if bg_params.adaptive_radius % 2 == 0: # sk >0.12.0 check for even value
bg_params.adaptive_radius += 1
bg_params
[13]:
BgParams(kind='li_adaptive', perc=0.1, radius=10, adaptive_radius=257, arcsinh_perc=80, erosion_disk=0, clip=False)
[14]:
bg_params.clip = False
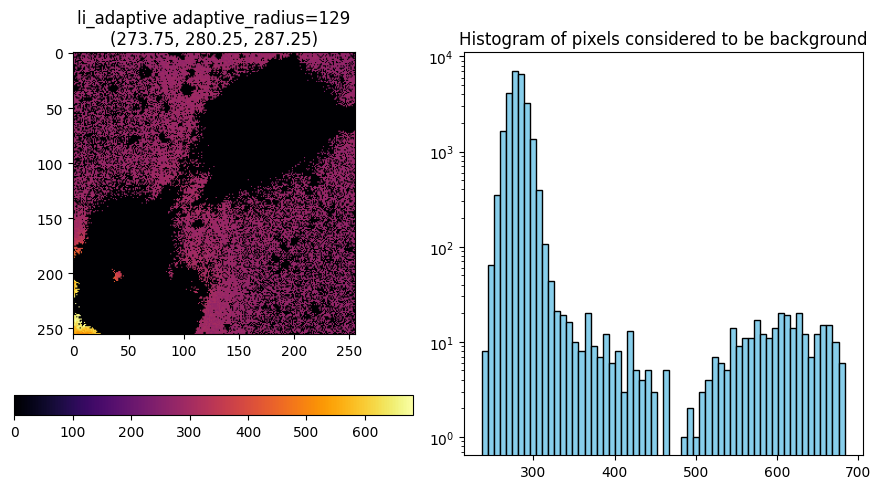
m, title, lim = segmentation._bg_li_adaptive(im, bg_params=bg_params)
title, lim
[14]:
('li_adaptive adaptive_radius=257', None)
[15]:
scipy.stats.distributions.norm.fit(im[m])
[15]:
(np.float64(278.5552310855508), np.float64(41.05308082242734))
[16]:
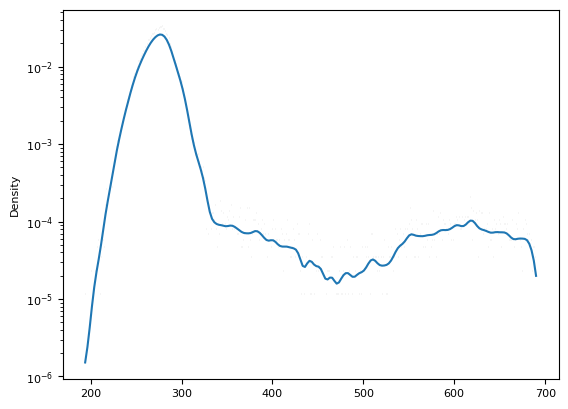
sns.histplot(im[m], kde=True, stat="density", log_scale=(False, True))
[16]:
<Axes: ylabel='Density'>
[17]:
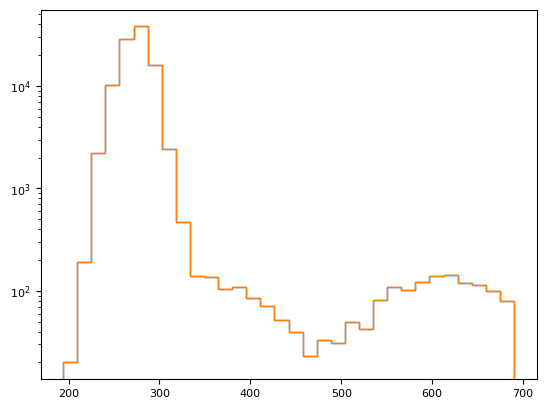
fig = plt.hist(im[m], histtype="step", bins=32, log=1)
fig = plt.hist(im[m], histtype="step", bins=32, log=1)
[18]:
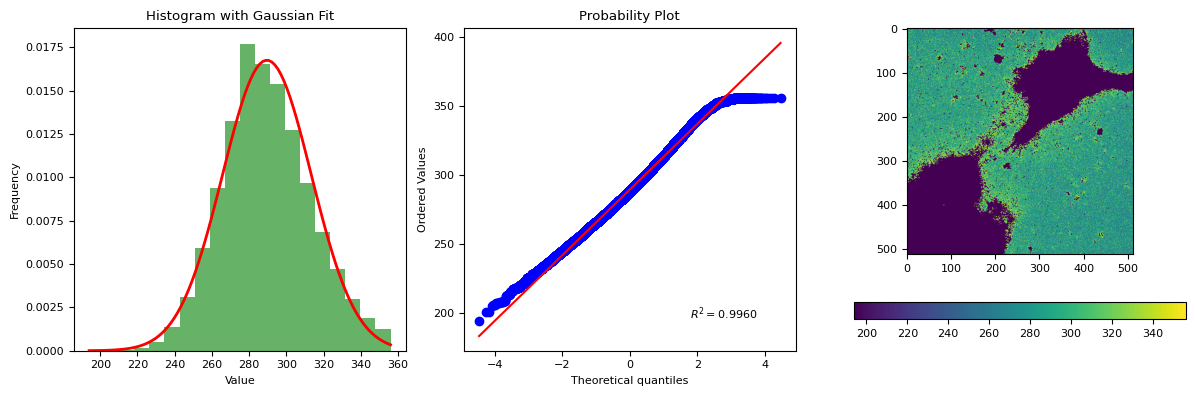
segmentation.calculate_bg_iteratively(im, probplot=True)
289.55334184900715 23.81726825652639
356
[18]:
BgResult(bg=np.float64(289.55334184900715), sd=np.float64(23.81726825652639), iqr=array([273., 288., 305.]), figures=[<Figure size 1200x400 with 4 Axes>])
1.2. background, AF, target cells#
[19]:
fp_mdtest = "/home/dati/GBM_persson/data/15.02.05_cal-GBM5-pBJclop/ph633/1_20_40.tif"
fp_mdtest = "/home/dati/dt-clop3/data/ionofori assay pH6.8 22.05.05/Clop FLATxy Gexp20ms Rexp3ms.tf8"
[20]:
nima.read_tiff(fp_mdtest, ["G", "C", "R"])
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/2636126201.py in ?()
----> 1 nima.read_tiff(fp_mdtest, ["G", "C", "R"])
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
70 (3, 4)
71
72 """
73 n_channels = len(channels)
---> 74 with TiffFile(fp) as tif:
75 im = tif.asarray()
76 axes = tif.series[0].axes
77 idx = axes.rfind("T")
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/ionofori assay pH6.8 22.05.05/Clop FLATxy Gexp20ms Rexp3ms.tf8'
[21]:
dim, md = nima.read_tiffmd(fp_mdtest, channels)
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/3787057078.py in ?()
----> 1 dim, md = nima.read_tiffmd(fp_mdtest, channels)
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
1072 def read_tiffmd(fp: Path, channels: Sequence[str]) -> tuple[Array, Metadata]:
1073 """Read multichannel TIFF timelapse image."""
1074 n_channels = len(channels)
-> 1075 rdr = TiffReader(fp)
1076 dim = da.from_zarr(rdr.aszarr()) # type: ignore[no-untyped-call]
1077 md = Metadata(rdr)
1078 if md.size_c[0] % n_channels:
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/ionofori assay pH6.8 22.05.05/Clop FLATxy Gexp20ms Rexp3ms.tf8'
[22]:
md.stage_position
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[22], line 1
----> 1 md.stage_position
NameError: name 'md' is not defined
[23]:
im = dim[:, 0].mean(axis=0).compute()
plt.imshow(im)
plt.colorbar()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[23], line 1
----> 1 im = dim[:, 0].mean(axis=0).compute()
2 plt.imshow(im)
3 plt.colorbar()
NameError: name 'dim' is not defined
[24]:
flat_im = scipy.ndimage.gaussian_filter(im, 150)
plt.imshow(flat_im)
plt.colorbar()
[24]:
<matplotlib.colorbar.Colorbar at 0x7f5af6f69a90>
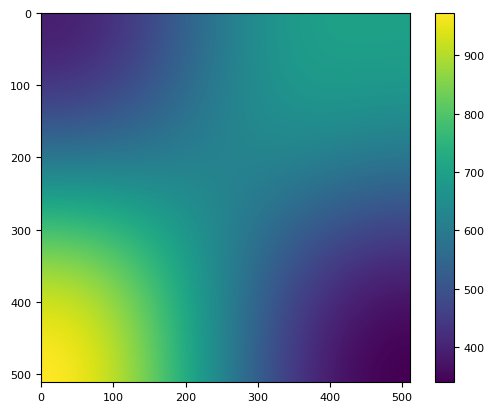
[25]:
plt.plot(flat_im[1060, :])
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
Cell In[25], line 1
----> 1 plt.plot(flat_im[1060, :])
IndexError: index 1060 is out of bounds for axis 0 with size 512
[26]:
fpm = "/home/dan/workspace/nima_io/tests/data/t4_1.tif"
[27]:
nima.read_tiffmd(fpm, ["G", "R2", "R", "C"])
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/3851534406.py in ?()
----> 1 nima.read_tiffmd(fpm, ["G", "R2", "R", "C"])
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
1072 def read_tiffmd(fp: Path, channels: Sequence[str]) -> tuple[Array, Metadata]:
1073 """Read multichannel TIFF timelapse image."""
1074 n_channels = len(channels)
-> 1075 rdr = TiffReader(fp)
1076 dim = da.from_zarr(rdr.aszarr()) # type: ignore[no-untyped-call]
1077 md = Metadata(rdr)
1078 if md.size_c[0] % n_channels:
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dan/workspace/nima_io/tests/data/t4_1.tif'
[28]:
nima.read_tiffmd(fpm, ["G", "R2", "R", "C"])
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/3851534406.py in ?()
----> 1 nima.read_tiffmd(fpm, ["G", "R2", "R", "C"])
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
1072 def read_tiffmd(fp: Path, channels: Sequence[str]) -> tuple[Array, Metadata]:
1073 """Read multichannel TIFF timelapse image."""
1074 n_channels = len(channels)
-> 1075 rdr = TiffReader(fp)
1076 dim = da.from_zarr(rdr.aszarr()) # type: ignore[no-untyped-call]
1077 md = Metadata(rdr)
1078 if md.size_c[0] % n_channels:
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dan/workspace/nima_io/tests/data/t4_1.tif'
[29]:
from nima import utils
[30]:
bgd = {}
for i, c in enumerate(channels):
bgd[c] = [
segmentation.calculate_bg_iteratively(dim[t, i].compute())
for t in range(md.size_t[0])
]
bgd
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[30], line 5
1 bgd = {}
2 for i, c in enumerate(channels):
3 bgd[c] = [
4 segmentation.calculate_bg_iteratively(dim[t, i].compute())
----> 5 for t in range(md.size_t[0])
6 ]
8 bgd
NameError: name 'md' is not defined
[31]:
for t in range(md.size_t[0]):
dim[t, 0] -= bgd[channels[0]][t].bg
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[31], line 1
----> 1 for t in range(md.size_t[0]):
2 dim[t, 0] -= bgd[channels[0]][t].bg
NameError: name 'md' is not defined
[32]:
plt.imshow(dim[0, 0])
plt.colorbar()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[32], line 1
----> 1 plt.imshow(dim[0, 0])
2 plt.colorbar()
NameError: name 'dim' is not defined
[33]:
segmentation.calculate_bg_iteratively(dim[0, 0].compute(), probplot=1)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[33], line 1
----> 1 segmentation.calculate_bg_iteratively(dim[0, 0].compute(), probplot=1)
NameError: name 'dim' is not defined
[34]:
bgd[channels[0]][1].bg
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[34], line 1
----> 1 bgd[channels[0]][1].bg
KeyError: 'G'
[35]:
t = 0
plt.imshow(dim[t, 0] - bgd[channels[0]][t].bg)
plt.colorbar()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[35], line 3
1 t = 0
----> 3 plt.imshow(dim[t, 0] - bgd[channels[0]][t].bg)
4 plt.colorbar()
NameError: name 'dim' is not defined
[36]:
m = segmentation.prob(dim[3, 0].compute(), 0, bgd[channels[0]][3].sd * 13) > 0.001
plt.imshow(~m)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[36], line 1
----> 1 m = segmentation.prob(dim[3, 0].compute(), 0, bgd[channels[0]][3].sd * 13) > 0.001
2 plt.imshow(~m)
NameError: name 'dim' is not defined
[37]:
asizeof(dim) / 1024**2
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[37], line 1
----> 1 asizeof(dim) / 1024**2
NameError: name 'dim' is not defined
[38]:
fpm = "/home/dan/workspace/nima_io/tests/data/t4_1.tif"
tifffile.TiffFile(fpm).series
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/1807629613.py in ?()
1 fpm = "/home/dan/workspace/nima_io/tests/data/t4_1.tif"
----> 2 tifffile.TiffFile(fpm).series
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dan/workspace/nima_io/tests/data/t4_1.tif'
[39]:
md = nima.Metadata(tifffile.TiffReader(fpm))
md
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/1703928088.py in ?()
----> 1 md = nima.Metadata(tifffile.TiffReader(fpm))
2 md
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dan/workspace/nima_io/tests/data/t4_1.tif'
[40]:
nima.read_tiffmd(fpm, ["G", "R2", "C", "R"])
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/143293133.py in ?()
----> 1 nima.read_tiffmd(fpm, ["G", "R2", "C", "R"])
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
1072 def read_tiffmd(fp: Path, channels: Sequence[str]) -> tuple[Array, Metadata]:
1073 """Read multichannel TIFF timelapse image."""
1074 n_channels = len(channels)
-> 1075 rdr = TiffReader(fp)
1076 dim = da.from_zarr(rdr.aszarr()) # type: ignore[no-untyped-call]
1077 md = Metadata(rdr)
1078 if md.size_c[0] % n_channels:
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dan/workspace/nima_io/tests/data/t4_1.tif'
[41]:
asizeof(dim) / 1024**2
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[41], line 1
----> 1 asizeof(dim) / 1024**2
NameError: name 'dim' is not defined
[42]:
bgr = segmentation.calculate_bg_iteratively(dim[1, 0].compute())
bgr.bg
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[42], line 1
----> 1 bgr = segmentation.calculate_bg_iteratively(dim[1, 0].compute())
2 bgr.bg
NameError: name 'dim' is not defined
[ ]:
[43]:
dim, md = nima.read_tiffmd(fp_mdtest, channels=["G", "R2", "C", "R"])
md
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/2107487587.py in ?()
----> 1 dim, md = nima.read_tiffmd(fp_mdtest, channels=["G", "R2", "C", "R"])
2
3 md
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
1072 def read_tiffmd(fp: Path, channels: Sequence[str]) -> tuple[Array, Metadata]:
1073 """Read multichannel TIFF timelapse image."""
1074 n_channels = len(channels)
-> 1075 rdr = TiffReader(fp)
1076 dim = da.from_zarr(rdr.aszarr()) # type: ignore[no-untyped-call]
1077 md = Metadata(rdr)
1078 if md.size_c[0] % n_channels:
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/ionofori assay pH6.8 22.05.05/Clop FLATxy Gexp20ms Rexp3ms.tf8'
[44]:
channels3 = ["G", "R", "C", "G2"]
dim3, _, _ = nima.read_tiff(fp_mdtest, channels3)
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/3014772201.py in ?()
1 channels3 = ["G", "R", "C", "G2"]
----> 2 dim3, _, _ = nima.read_tiff(fp_mdtest, channels3)
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
70 (3, 4)
71
72 """
73 n_channels = len(channels)
---> 74 with TiffFile(fp) as tif:
75 im = tif.asarray()
76 axes = tif.series[0].axes
77 idx = axes.rfind("T")
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/ionofori assay pH6.8 22.05.05/Clop FLATxy Gexp20ms Rexp3ms.tf8'
[45]:
dim3, bgv3, bgf3 = nima.d_bg(
dim3, segmentation.BgParams(kind="li_adaptive"), downscale=(2, 2)
)
c = dim3["C"]
g = dim3["G"]
r = dim3["R"]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[45], line 2
1 dim3, bgv3, bgf3 = nima.d_bg(
----> 2 dim3, segmentation.BgParams(kind="li_adaptive"), downscale=(2, 2)
3 )
4 c = dim3["C"]
5 g = dim3["G"]
NameError: name 'dim3' is not defined
[46]:
bc = bgv3["C"]
bg = bgv3["G"][3]
br = bgv3["R"][3]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[46], line 1
----> 1 bc = bgv3["C"]
2 bg = bgv3["G"][3]
3 br = bgv3["R"][3]
NameError: name 'bgv3' is not defined
[47]:
skimage.io.imshow(dim3["R"][1])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[47], line 1
----> 1 skimage.io.imshow(dim3["R"][1])
NameError: name 'dim3' is not defined
[48]:
plt.plot(dim3["R"][1][80, 30:])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[48], line 1
----> 1 plt.plot(dim3["R"][1][80, 30:])
NameError: name 'dim3' is not defined
[49]:
green = dim[0].compute()
bgg = segmentation.calculate_bg_iteratively(green)
bgg
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[49], line 1
----> 1 green = dim[0].compute()
2 bgg = segmentation.calculate_bg_iteratively(green)
3 bgg
NameError: name 'dim' is not defined
[50]:
mask = segmentation.prob(green, bgg.bg, bgg.sd) > 0.01
green[mask]
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[50], line 1
----> 1 mask = segmentation.prob(green, bgg.bg, bgg.sd) > 0.01
2 green[mask]
NameError: name 'green' is not defined
[51]:
cyan = dim[2].compute()
bgc = segmentation.calculate_bg_iteratively(cyan)
mask = segmentation.prob(cyan, bgc.bg, bgc.sd) < 0.00000000000000000000001
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[51], line 1
----> 1 cyan = dim[2].compute()
2 bgc = segmentation.calculate_bg_iteratively(cyan)
3 mask = segmentation.prob(cyan, bgc.bg, bgc.sd) < 0.00000000000000000000001
NameError: name 'dim' is not defined
[52]:
red = dim[3].compute()
red2 = dim[1].compute()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[52], line 1
----> 1 red = dim[3].compute()
2 red2 = dim[1].compute()
NameError: name 'dim' is not defined
[53]:
plt.figure(figsize=(7.5, 7.5))
plt.subplot(3, 2, 1)
plt.hexbin(green[mask], cyan[mask], bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("green")
plt.ylabel("cyan")
plt.subplot(3, 2, 2)
plt.hexbin(red[mask], cyan[mask], bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("red")
plt.ylabel("cyan")
ax = plt.subplot(3, 2, 4)
plt.hexbin(red[mask], green[mask], bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("red")
plt.ylabel("green")
ax = plt.subplot(3, 2, 5)
plt.hexbin(red2[mask], green[mask], bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("red2")
plt.ylabel("green")
ax = plt.subplot(3, 2, 6)
plt.hexbin(red2[mask], cyan[mask], bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("red2")
plt.ylabel("cyan")
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[53], line 4
1 plt.figure(figsize=(7.5, 7.5))
3 plt.subplot(3, 2, 1)
----> 4 plt.hexbin(green[mask], cyan[mask], bins="log", cmap=plt.cm.viridis_r)
5 cb = plt.colorbar()
6 plt.xlabel("green")
NameError: name 'green' is not defined
[54]:
from mpl_toolkits.axes_grid1.inset_locator import mark_inset
plt.figure(figsize=(10, 8))
plt.subplot(2, 2, 1)
plt.hexbin(g.ravel(), c.ravel(), bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("green")
plt.ylabel("cyan")
plt.subplot(2, 2, 2)
plt.hexbin(r.ravel(), c.ravel(), bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("red")
plt.ylabel("cyan")
ax = plt.subplot(2, 2, 4)
plt.hexbin(r.ravel(), g.ravel(), bins="log", cmap=plt.cm.viridis_r)
cb = plt.colorbar()
plt.xlabel("red")
plt.ylabel("green")
axins = plt.axes([0.2, 0.12, 0.28, 0.28])
axins.hexbin(
r.ravel(), g.ravel(), extent=(0, 80, 0, 150), bins="log", cmap=plt.cm.viridis_r
)
mark_inset(ax, axins, loc1=1, loc2=4, fc="none", ec="0.5")
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[54], line 5
3 plt.figure(figsize=(10, 8))
4 plt.subplot(2, 2, 1)
----> 5 plt.hexbin(g.ravel(), c.ravel(), bins="log", cmap=plt.cm.viridis_r)
6 cb = plt.colorbar()
7 plt.xlabel("green")
NameError: name 'g' is not defined
[55]:
import statsmodels.api as sm
f = sm.qqplot_2samples(red[~mask], green[~mask])
f = sm.qqplot_2samples(red[~mask], cyan[~mask])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[55], line 3
1 import statsmodels.api as sm
----> 3 f = sm.qqplot_2samples(red[~mask], green[~mask])
4 f = sm.qqplot_2samples(red[~mask], cyan[~mask])
NameError: name 'red' is not defined
1.3. flat image correction#
[56]:
g1a = nima.read_tiff("/home/dati/dt-clop3/data/210917/3alpha/c1_2.tf8", channels)[0][
"G"
][2]
g2a = nima.read_tiff("/home/dati/dt-clop3/data/210917/3alpha/c2_2.tf8", channels)[0][
"G"
][2]
g3a = nima.read_tiff("/home/dati/dt-clop3/data/210917/3alpha/c3_2.tf8", channels)[0][
"G"
][2]
g1 = nima.read_tiff("/home/dati/dt-clop3/data/210917/3/c1_2.tf8", channels)[0]["G"][2]
g2 = nima.read_tiff("/home/dati/dt-clop3/data/210917/3/c2_2.tf8", channels)[0]["G"][2]
g3 = nima.read_tiff("/home/dati/dt-clop3/data/210917/3/c3_2.tf8", channels)[0]["G"][2]
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/4145832345.py in ?()
----> 1 g1a = nima.read_tiff("/home/dati/dt-clop3/data/210917/3alpha/c1_2.tf8", channels)[0][
2 "G"
3 ][2]
4 g2a = nima.read_tiff("/home/dati/dt-clop3/data/210917/3alpha/c2_2.tf8", channels)[0][
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
70 (3, 4)
71
72 """
73 n_channels = len(channels)
---> 74 with TiffFile(fp) as tif:
75 im = tif.asarray()
76 axes = tif.series[0].axes
77 idx = axes.rfind("T")
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/210917/3alpha/c1_2.tf8'
[57]:
plt.subplot(1, 2, 1)
skimage.io.imshow(
skimage.filters.gaussian(np.max([g1, g2, g3, g1a, g2a, g3a], axis=0), sigma=500)
)
plt.subplot(1, 2, 2)
skimage.io.imshow(skimage.filters.gaussian(np.max([g1, g2, g3], axis=0), sigma=500))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[57], line 3
1 plt.subplot(1, 2, 1)
2 skimage.io.imshow(
----> 3 skimage.filters.gaussian(np.max([g1, g2, g3, g1a, g2a, g3a], axis=0), sigma=500)
4 )
5 plt.subplot(1, 2, 2)
6 skimage.io.imshow(skimage.filters.gaussian(np.max([g1, g2, g3], axis=0), sigma=500))
NameError: name 'g1' is not defined
1.4. example w/out @Gain#
[58]:
fp24 = "/home/dati/dt-clop3/data/210917/3/c1_2.tf8"
[59]:
dim, md = nima.read_tiffmd(fp24, channels=["G", "R", "C"])
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_2665/2668091231.py in ?()
----> 1 dim, md = nima.read_tiffmd(fp24, channels=["G", "R", "C"])
~/work/nima/nima/src/nima/nima.py in ?(fp, channels)
1072 def read_tiffmd(fp: Path, channels: Sequence[str]) -> tuple[Array, Metadata]:
1073 """Read multichannel TIFF timelapse image."""
1074 n_channels = len(channels)
-> 1075 rdr = TiffReader(fp)
1076 dim = da.from_zarr(rdr.aszarr()) # type: ignore[no-untyped-call]
1077 md = Metadata(rdr)
1078 if md.size_c[0] % n_channels:
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size, omexml, superres, _multifile, _useframes, _parent, **is_flags)
4260 raise ValueError('invalid OME-XML')
4261 self._omexml = omexml
4262 self.is_ome = True
4263
-> 4264 fh = FileHandle(file, mode=mode, name=name, offset=offset, size=size)
4265 self._fh = fh
4266 self._multifile = True if _multifile is None else bool(_multifile)
4267 self._files = {fh.name: self}
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self, file, mode, name, offset, size)
13440 self._offset = -1 if offset is None else offset
13441 self._size = -1 if size is None else size
13442 self._close = True
13443 self._lock = NullContext()
> 13444 self.open()
13445 assert self._fh is not None
~/work/nima/nima/.venv/lib/python3.13/site-packages/tifffile/tifffile.py in ?(self)
13459 if self._mode not in {'rb', 'r+b', 'wb', 'xb'}:
13460 raise ValueError(f'invalid mode {self._mode}')
13461 self._file = os.path.realpath(self._file)
13462 self._dir, self._name = os.path.split(self._file)
> 13463 self._fh = open(self._file, self._mode, encoding=None)
13464 self._close = True
13465 self._offset = max(0, self._offset)
13466 elif isinstance(self._file, FileHandle):
FileNotFoundError: [Errno 2] No such file or directory: '/home/dati/dt-clop3/data/210917/3/c1_2.tf8'
[60]:
sns.histplot(dim[8, 0].ravel())
plt.yscale("log")
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[60], line 1
----> 1 sns.histplot(dim[8, 0].ravel())
2 plt.yscale("log")
NameError: name 'dim' is not defined
[61]:
utils.bg(dim[8, 0].compute())
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[61], line 1
----> 1 utils.bg(dim[8, 0].compute())
NameError: name 'dim' is not defined
[62]:
bgr = segmentation.calculate_bg_iteratively(dim[8, 1].compute())
bgr
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[62], line 1
----> 1 bgr = segmentation.calculate_bg_iteratively(dim[8, 1].compute())
2 bgr
NameError: name 'dim' is not defined
[63]:
sns.histplot(
dim[8, 1].compute()[
(segmentation.prob(dim[8, 1].compute(), bgr.bg, bgr.sd) > 0.001)
]
)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[63], line 2
1 sns.histplot(
----> 2 dim[8, 1].compute()[
3 (segmentation.prob(dim[8, 1].compute(), bgr.bg, bgr.sd) > 0.001)
4 ]
5 )
NameError: name 'dim' is not defined
[64]:
mask = (
segmentation.prob(dim[8, 1].compute(), bgr.bg, bgr.sd)
> 0.0000000000000000000000000000000000000000000000000000000000000000001
)
plt.imshow(~mask)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[64], line 2
1 mask = (
----> 2 segmentation.prob(dim[8, 1].compute(), bgr.bg, bgr.sd)
3 > 0.0000000000000000000000000000000000000000000000000000000000000000001
4 )
5 plt.imshow(~mask)
NameError: name 'dim' is not defined
1.5. BIAS and FLAT#
TODO:
pytest
build a function callable from both the library and the CLI