3. Getting started with prenspire#
import random
from pathlib import Path
import lmfit
import numpy as np
from clophfit import prenspire
from clophfit.fitting.core import (
DataArray,
Dataset,
analyze_spectra,
analyze_spectra_glob,
fit_binding_glob,
weight_multi_ds_titration,
)
from clophfit.fitting.plotting import plot_emcee
%load_ext autoreload
%autoreload 2
tpath = Path("../../tests/EnSpire")
ef1 = prenspire.EnspireFile(tpath / "h148g-spettroC.csv")
ef2 = prenspire.EnspireFile(tpath / "e2-T-without_sample_column.csv")
ef3 = prenspire.EnspireFile(tpath / "24well_clop0_95.csv")
ef3.wells, ef3._wells_platemap, ef3._platemap
(['A03', 'A04', 'A05', 'A06', 'B01', 'B02', 'C01', 'C02', 'C03'],
['A03', 'A04', 'A05', 'A06', 'B01', 'B02', 'C01', 'C02', 'C03'],
[['A', ' ', ' ', '- ', '- ', '- ', '- '],
['B', '- ', '- ', ' ', ' ', ' ', ' '],
['C', '- ', '- ', '- ', ' ', ' ', ' '],
['D', ' ', ' ', ' ', ' ', ' ', ' ']])
ef1.__dict__.keys()
dict_keys(['file', 'verbose', 'metadata', 'measurements', 'wells', '_ini', '_fin', '_wells_platemap', '_platemap'])
ef1.measurements.keys(), ef2.measurements.keys()
(dict_keys(['A']), dict_keys(['B', 'A', 'C', 'D', 'E', 'F', 'G', 'H']))
when testing each spectra for the presence of a single wavelength in the appropriate monochromator
ef2.measurements["A"]["metadata"]
{'temp': '25',
'Monochromator': 'Excitation',
'Min wavelength': '400',
'Max wavelength': '510',
'Wavelength': '530',
'Using of excitation filter': 'Top',
'Measurement height': '8.9',
'Number of flashes': '50',
'Number of flashes integrated': '50',
'Flash power': '100'}
ef2.measurements["A"].keys()
dict_keys(['metadata', 'lambda', 'F01', 'F02', 'F03', 'F04', 'F05', 'F06', 'F07'])
random.seed(11)
random.sample(ef1.measurements["A"]["F01"], 7)
[2163.0, 607.0, 1846.0, 517.0, 572.0, 2145.0, 2028.0]
fp = tpath / "h148g-spettroC-nota.csv"
n1 = prenspire.Note(fp, verbose=1)
n1._note.set_index("Well").loc["A01", ["Name", "Temp"]]
Wells ['A01', 'A02']...['G04', 'G05'] generated.
Name H148G
Temp 20.0
Name: A01, dtype: object
n1.__dict__.keys()
dict_keys(['fpath', 'verbose', 'wells', '_note', 'titrations'])
n1.wells == ef1.wells, n1.wells == ef2.wells
(True, False)
n1.build_titrations(ef1)
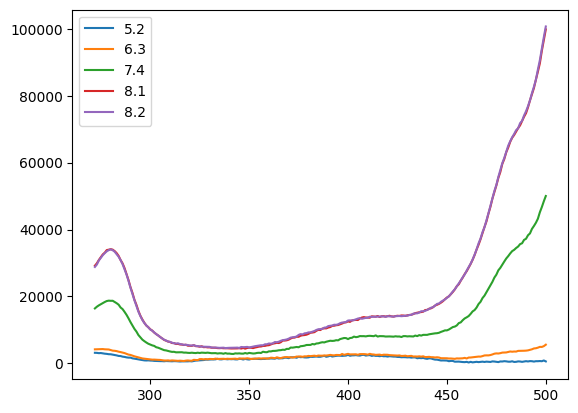
tit0 = n1.titrations["H148G"][20.0]["Cl_0.0"]["A"]
tit3 = n1.titrations["H148G"][20.0]["pH_7.4"]["A"]
tit0
| 5.2 | 6.3 | 7.4 | 8.1 | 8.2 | |
|---|---|---|---|---|---|
| 272.0 | 3151.0 | 4181.0 | 16413.0 | 29192.0 | 28816.0 |
| 273.0 | 3130.0 | 4204.0 | 16926.0 | 29909.0 | 29545.0 |
| 274.0 | 3043.0 | 4232.0 | 17331.0 | 30900.0 | 30750.0 |
| 275.0 | 3079.0 | 4283.0 | 17680.0 | 31717.0 | 31547.0 |
| 276.0 | 2975.0 | 4264.0 | 18020.0 | 32564.0 | 32336.0 |
| ... | ... | ... | ... | ... | ... |
| 496.0 | 636.0 | 4689.0 | 43230.0 | 87203.0 | 87842.0 |
| 497.0 | 683.0 | 4923.0 | 45173.0 | 89719.0 | 90666.0 |
| 498.0 | 632.0 | 4900.0 | 46725.0 | 93452.0 | 94101.0 |
| 499.0 | 854.0 | 5140.0 | 48452.0 | 96643.0 | 97506.0 |
| 500.0 | 573.0 | 5573.0 | 50025.0 | 99847.0 | 100715.0 |
229 rows × 5 columns
tit0.plot()
tit3.plot()
<Axes: >
ef = prenspire.EnspireFile(tpath / "G10.csv")
fp = tpath / "NTT-G10_note.csv"
nn = prenspire.Note(fp, verbose=1)
nn.build_titrations(ef)
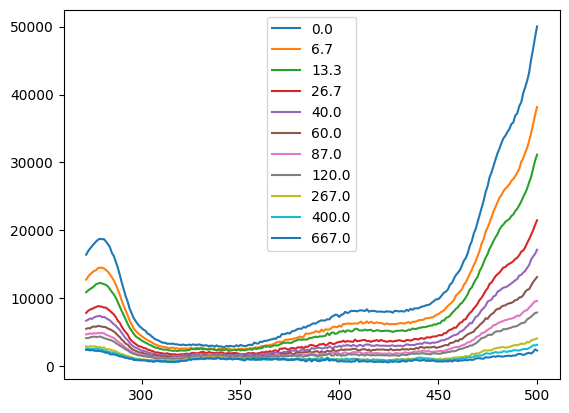
spectra = nn.titrations["NTT-G10"][20.0]["Cl_0.0"]["C"]
Wells ['D01', 'D02']...['G07', 'G08'] generated.
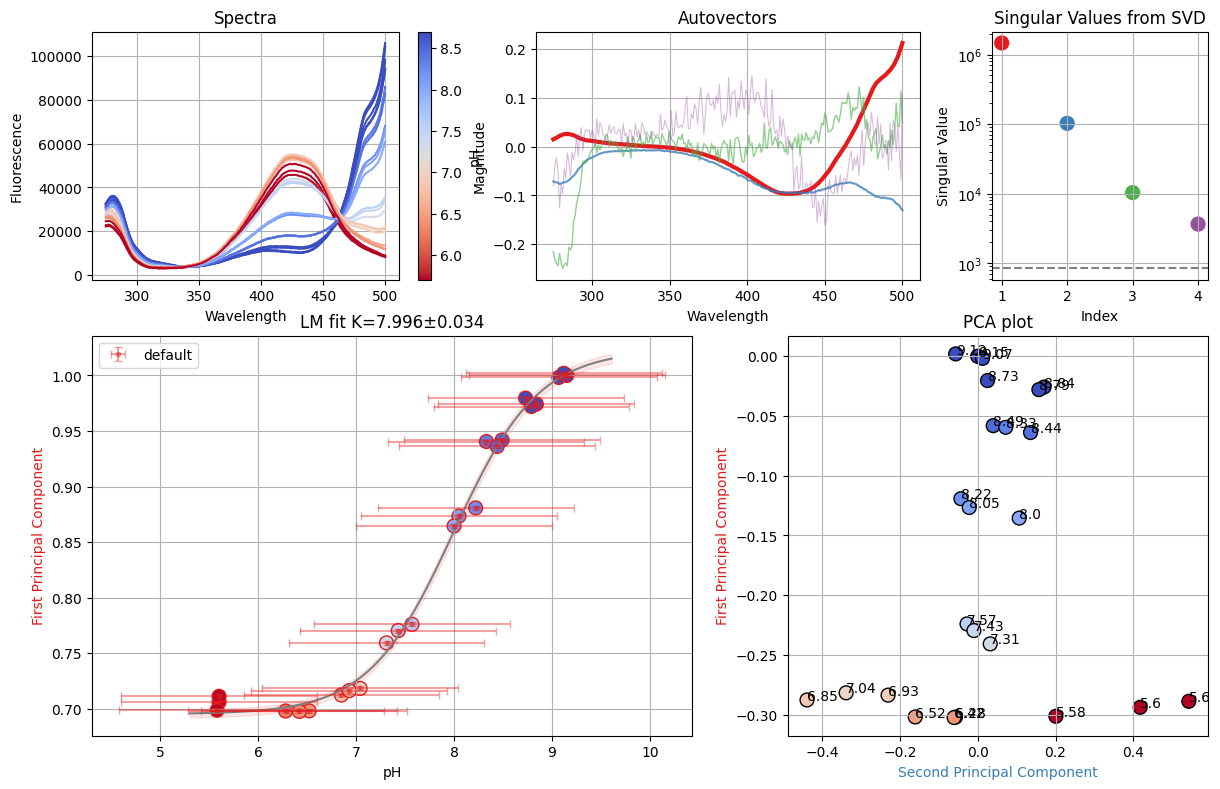
f_res = analyze_spectra(spectra, is_ph=True, band=None)
print(f_res.result.chisqr)
f_res.figure
551.9999999999978
def dataset_from_lres(lkey, lres, is_ph):
data = {}
for k, res in zip(lkey, lres, strict=False):
x = res.mini.userargs[0]["default"].x
y = res.mini.userargs[0]["default"].y
data[k] = DataArray(x, y)
return Dataset(data, is_ph=is_ph)
spectra_A = nn.titrations["NTT-G10"][20.0]["Cl_0.0"]["A"]
spectra_C = nn.titrations["NTT-G10"][20.0]["Cl_0.0"]["C"]
spectra_D = nn.titrations["NTT-G10"][20.0]["Cl_0.0"]["D"]
resA = analyze_spectra(spectra_A, is_ph=True, band=(466, 510))
resC = analyze_spectra(spectra_C, is_ph=True, band=(470, 500))
resD = analyze_spectra(spectra_D, is_ph=True, band=(450, 600))
ds_bands = dataset_from_lres(["A", "C", "D"], [resA, resC, resD], is_ph=True)
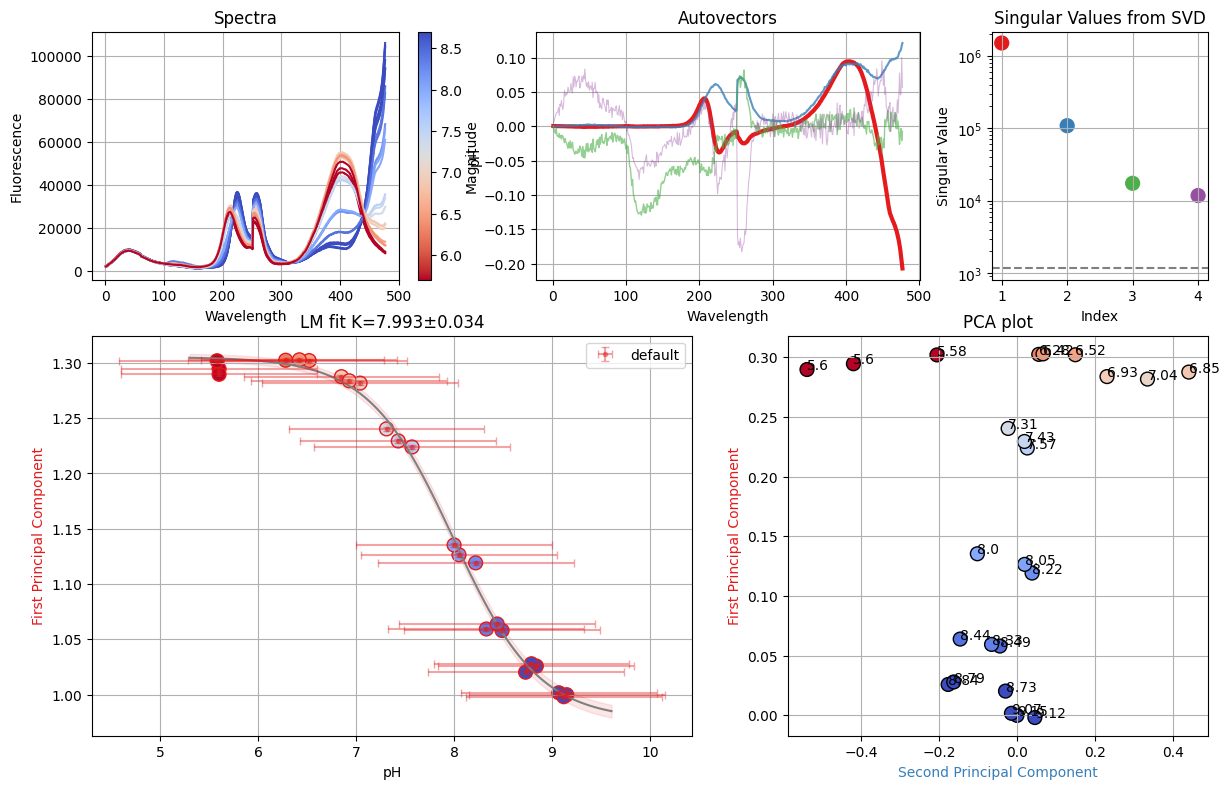
resA = analyze_spectra(spectra_A, is_ph=True)
resC = analyze_spectra(spectra_C, is_ph=True)
resD = analyze_spectra(spectra_D, is_ph=True)
ds_svd = dataset_from_lres(["A", "C", "D"], [resA, resC, resD], is_ph=True)
dbands = {"D": (466, 510)}
tit = nn.titrations["NTT-G10"][20.0]["Cl_0.0"]
sgres = analyze_spectra_glob(tit, ds_svd, dbands)
sgres.svd.figure
The chain is shorter than 50 times the integrated autocorrelation time for 1 parameter(s). Use this estimate with caution and run a longer chain!
N/50 = 108;
tau: [ 85.19893782 130.44068731 84.24663709 58.03314941 42.16063864]
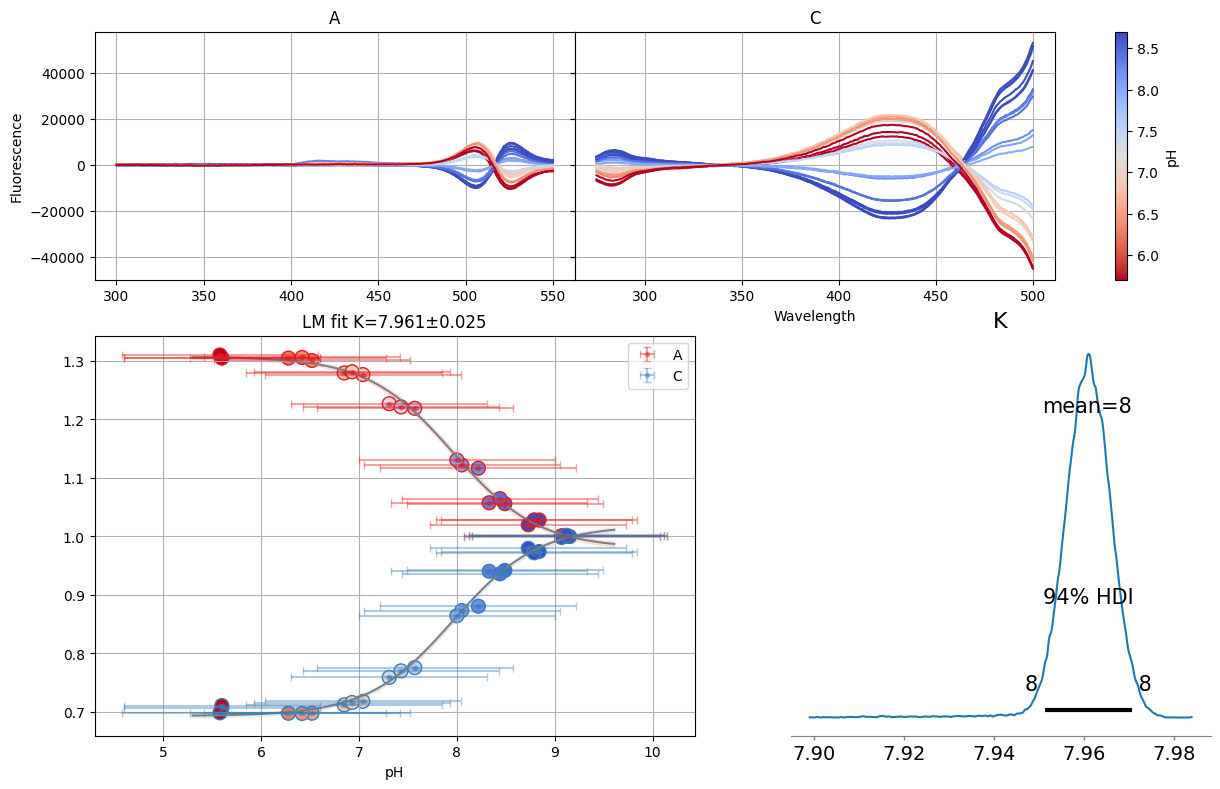
sgres.gsvd.figure
dbands = {"A": (466, 510), "C": (470, 500), "D": (450, 600)}
sgres = analyze_spectra_glob(nn.titrations["NTT-G10"][20.0]["Cl_0.0"], ds_bands, dbands)
sgres.bands.figure
The chain is shorter than 50 times the integrated autocorrelation time for 5 parameter(s). Use this estimate with caution and run a longer chain!
N/50 = 108;
tau: [245.25503923 285.78022353 54.0203079 245.68233136 76.78561808
251.85092135 255.61380209]
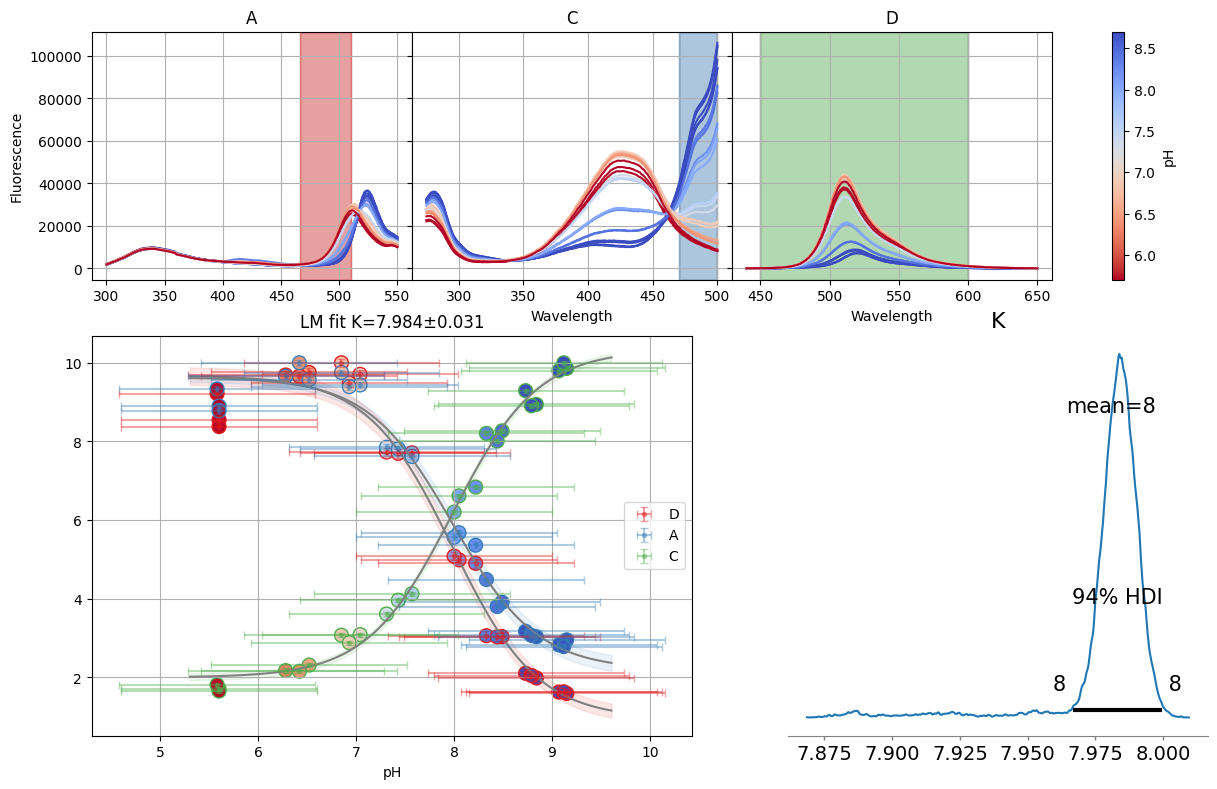
ci = lmfit.conf_interval(sgres.bands.mini, sgres.bands.result)
print(lmfit.ci_report(ci, ndigits=2, with_offset=False))
99.73% 95.45% 68.27% _BEST_ 68.27% 95.45% 99.73%
K : 7.89 7.92 7.95 7.98 8.02 8.05 8.08
S0_D: 0.21 0.47 0.72 0.96 1.19 1.42 1.67
S1_D: 9.12 9.31 9.50 9.68 9.85 10.04 10.23
S0_A: 1.65 1.84 2.02 2.20 2.37 2.54 2.72
S1_A: 9.23 9.37 9.50 9.63 9.76 9.89 10.03
S0_C: 9.90 10.05 10.19 10.33 10.47 10.62 10.79
S1_C: 1.70 1.81 1.91 2.00 2.10 2.19 2.29
weight_multi_ds_titration(ds_svd)
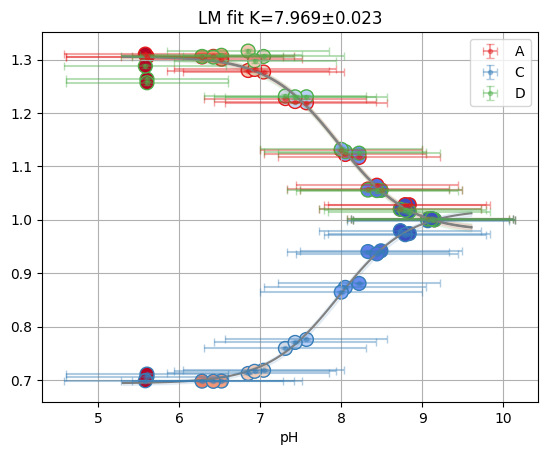
res = fit_binding_glob(ds_svd)
res.figure
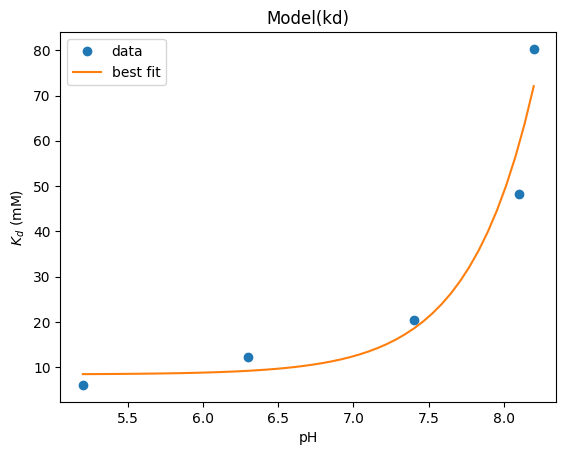
xx = np.array([5.2, 6.3, 7.4, 8.1, 8.2])
yy = np.array([6.05, 12.2, 20.38, 48.2, 80.3])
def kd(x, kd1, pka):
return kd1 * (1 + 10 ** (pka - x)) / 10 ** (pka - x)
model = lmfit.Model(kd)
params = lmfit.Parameters()
params.add("kd1", value=10.0)
params.add("pka", value=6.6)
result = model.fit(yy, params, x=xx)
result.plot_fit(numpoints=50, ylabel="$K_d$ (mM)", xlabel="pH")
<Axes: title={'center': 'Model(kd)'}, xlabel='pH', ylabel='$K_d$ (mM)'>
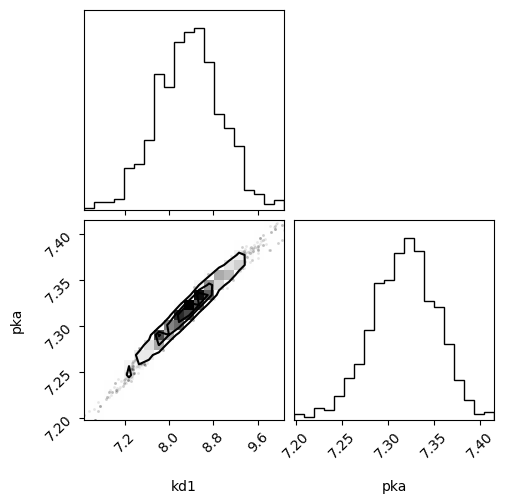
import os
result_emcee = result.emcee(
steps=int(os.environ.get("CLOPHFIT_DOCS_EMCEE_STEPS", "1800")),
burn=int(os.environ.get("CLOPHFIT_DOCS_EMCEE_BURN", "150")),
workers=int(os.environ.get("CLOPHFIT_EMCEE_WORKERS", "1")),
nwalkers=int(os.environ.get("CLOPHFIT_DOCS_EMCEE_NWALKERS", "10")),
seed=1,
progress=False,
)
fig = plot_emcee(result_emcee.flatchain)
The chain is shorter than 50 times the integrated autocorrelation time for 2 parameter(s). Use this estimate with caution and run a longer chain!
N/50 = 6;
tau: [21.09140903 21.25484981]
import pymc as pm
# Assuming x_data and y_data are your data
with pm.Model() as model:
kd1 = pm.Normal(
"kd1", mu=result.params["kd1"].value, sigma=result.params["kd1"].stderr
)
pka = pm.Normal(
"pka", mu=result.params["pka"].value, sigma=result.params["pka"].stderr
)
y_pred = pm.Deterministic("y_pred", kd(xx, kd1, pka))
likelihood = pm.Normal("y", mu=y_pred, sigma=1, observed=yy)
trace = pm.sample(1000, tune=2000, random_seed=1, chains=8, cores=8)
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (8 chains in 8 jobs)
NUTS: [kd1, pka]
/home/runner/work/ClopHfit/ClopHfit/.venv/lib/python3.14/site-packages/pymc/step_methods/hmc/quadpotential.py:316: RuntimeWarning: overflow encountered in dot
return 0.5 * np.dot(x, v_out)
/home/runner/work/ClopHfit/ClopHfit/.venv/lib/python3.14/site-packages/pymc/step_methods/hmc/quadpotential.py:316: RuntimeWarning: overflow encountered in dot
return 0.5 * np.dot(x, v_out)
Sampling 8 chains for 2_000 tune and 1_000 draw iterations (16_000 + 8_000 draws total) took 28 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
pm.model_to_graphviz(model)
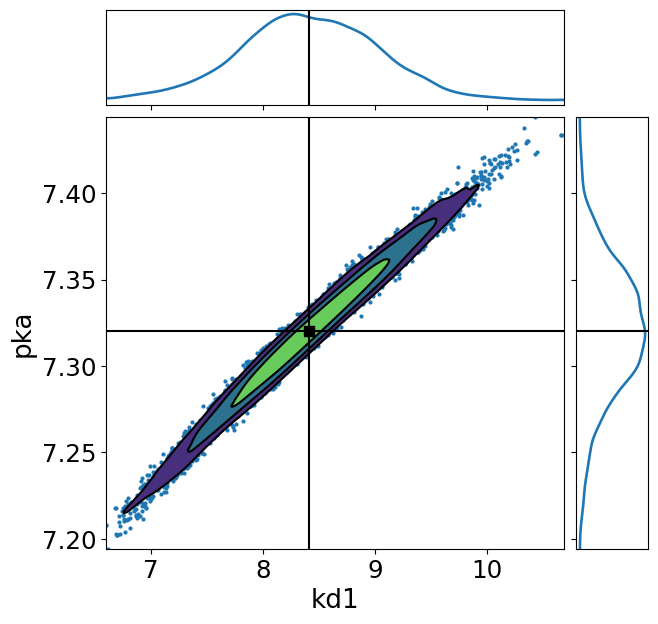
import arviz as az
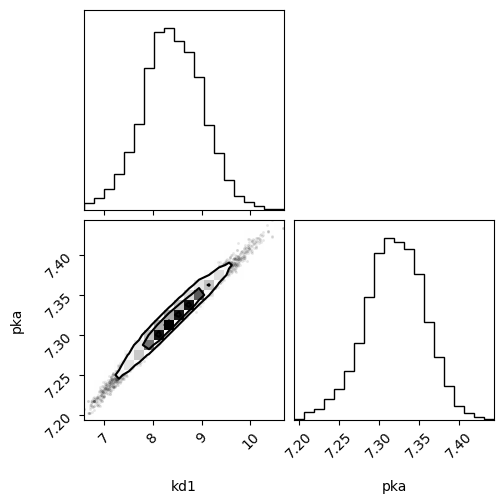
ax = az.plot_pair(
trace,
divergences=1,
var_names=["kd1", "pka"],
kind=["kde", "scatter"],
kde_kwargs={"fill_last": False},
marginals=True,
point_estimate="mean",
figsize=(7, 7),
)
import corner
# Extract the posterior samples for the parameters of interest
kd1_samples = trace.posterior["kd1"].to_numpy().flatten()
pka_samples = trace.posterior["pka"].to_numpy().flatten()
# Ensure the samples are in the correct format for the corner plotpm.plot_posterior(trace, var_names=["R0", "R1", "K"])
samples_array = np.column_stack([kd1_samples, pka_samples])
# Plot the corner plot
f = corner.corner(samples_array, labels=["kd1", "pka"])
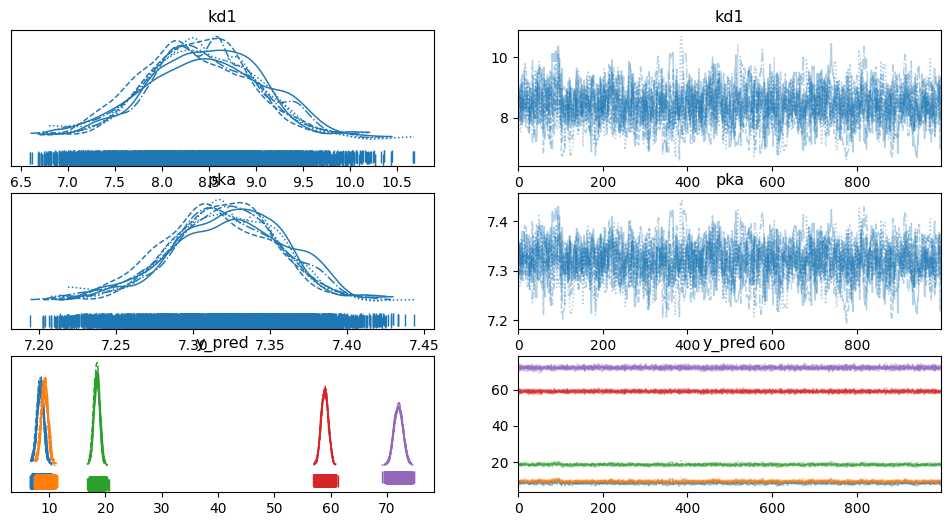
pm.plot_trace(trace, rug=True)
array([[<Axes: title={'center': 'kd1'}>, <Axes: title={'center': 'kd1'}>],
[<Axes: title={'center': 'pka'}>, <Axes: title={'center': 'pka'}>],
[<Axes: title={'center': 'y_pred'}>,
<Axes: title={'center': 'y_pred'}>]], dtype=object)
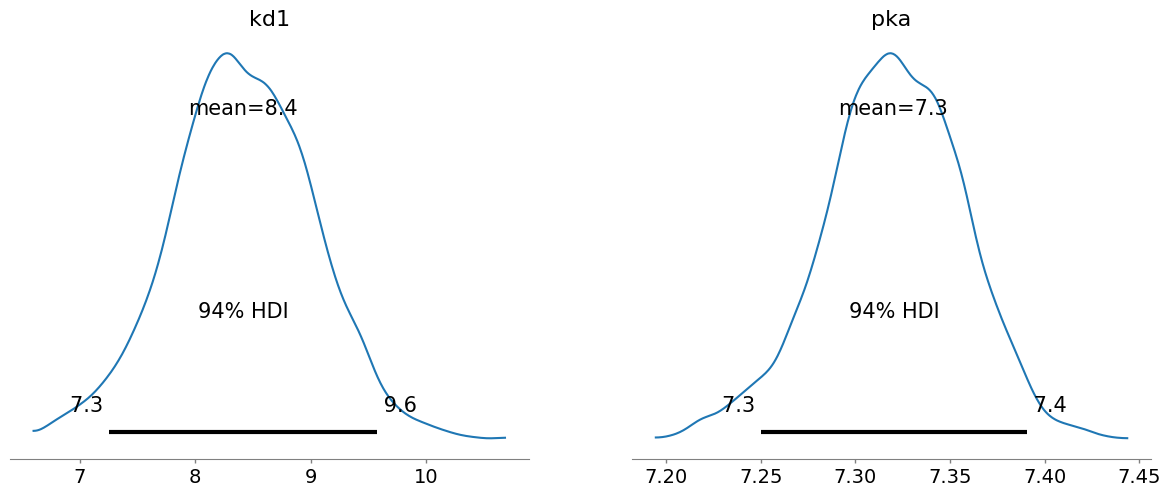
pm.plot_posterior(trace, var_names=["kd1", "pka"])
array([<Axes: title={'center': 'kd1'}>, <Axes: title={'center': 'pka'}>],
dtype=object)
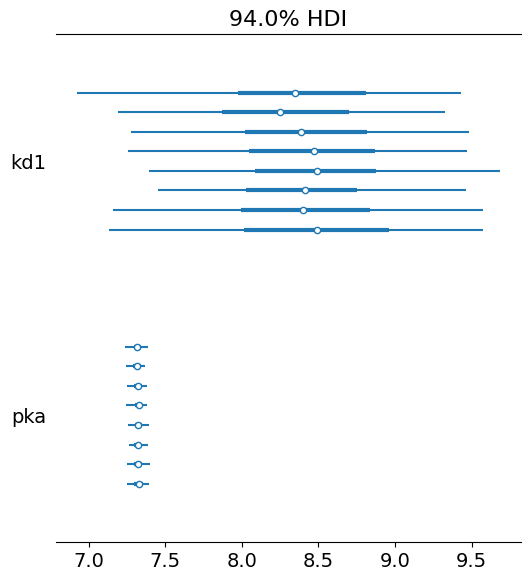
pm.plot_forest(trace, var_names=["kd1", "pka"])
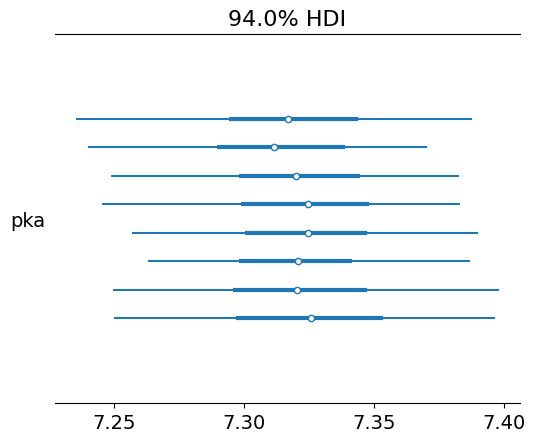
pm.plot_forest(trace, var_names=["pka"])
array([<Axes: title={'center': '94.0% HDI'}>], dtype=object)